Extended blood stage sensitivity profiles of Plasmodium cynomolgi to doxycycline and tafenoquine, as a model for Plasmodium vivax
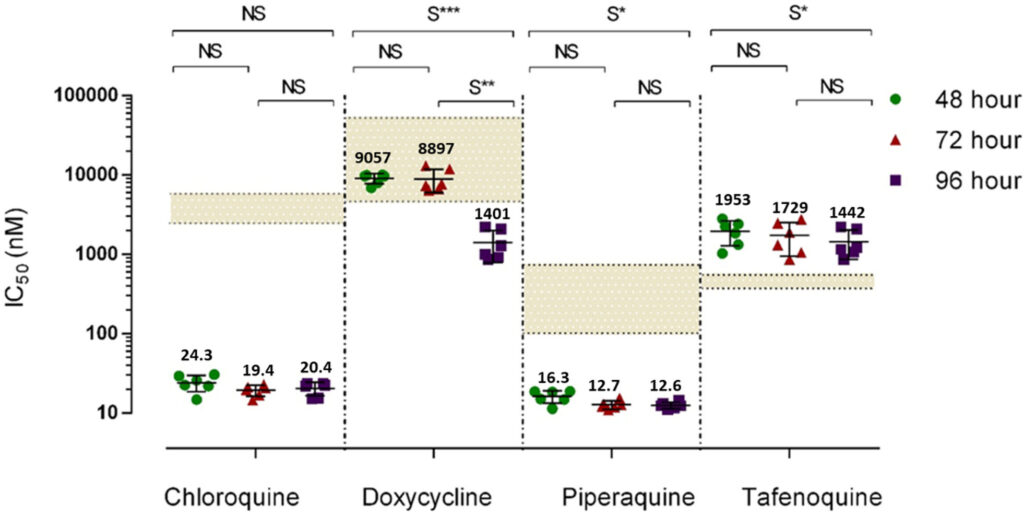
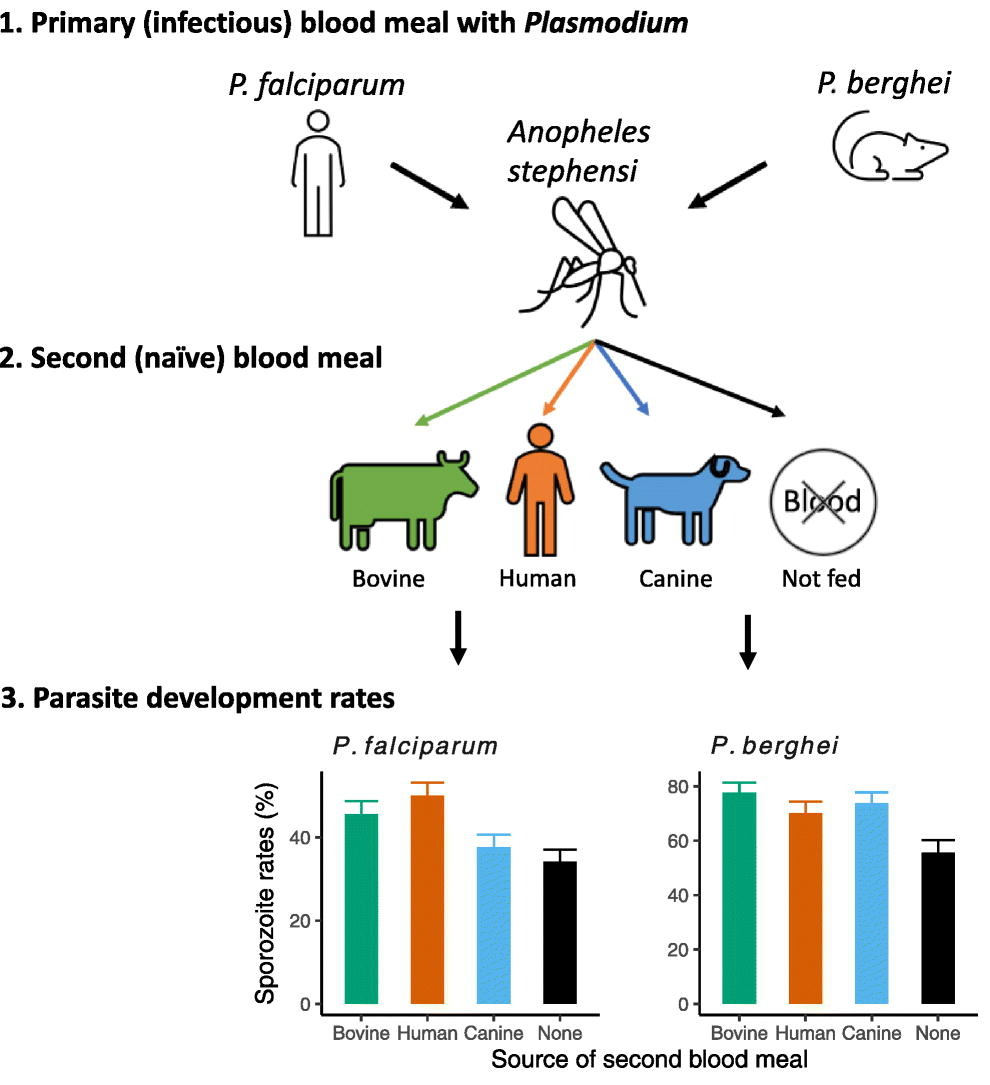
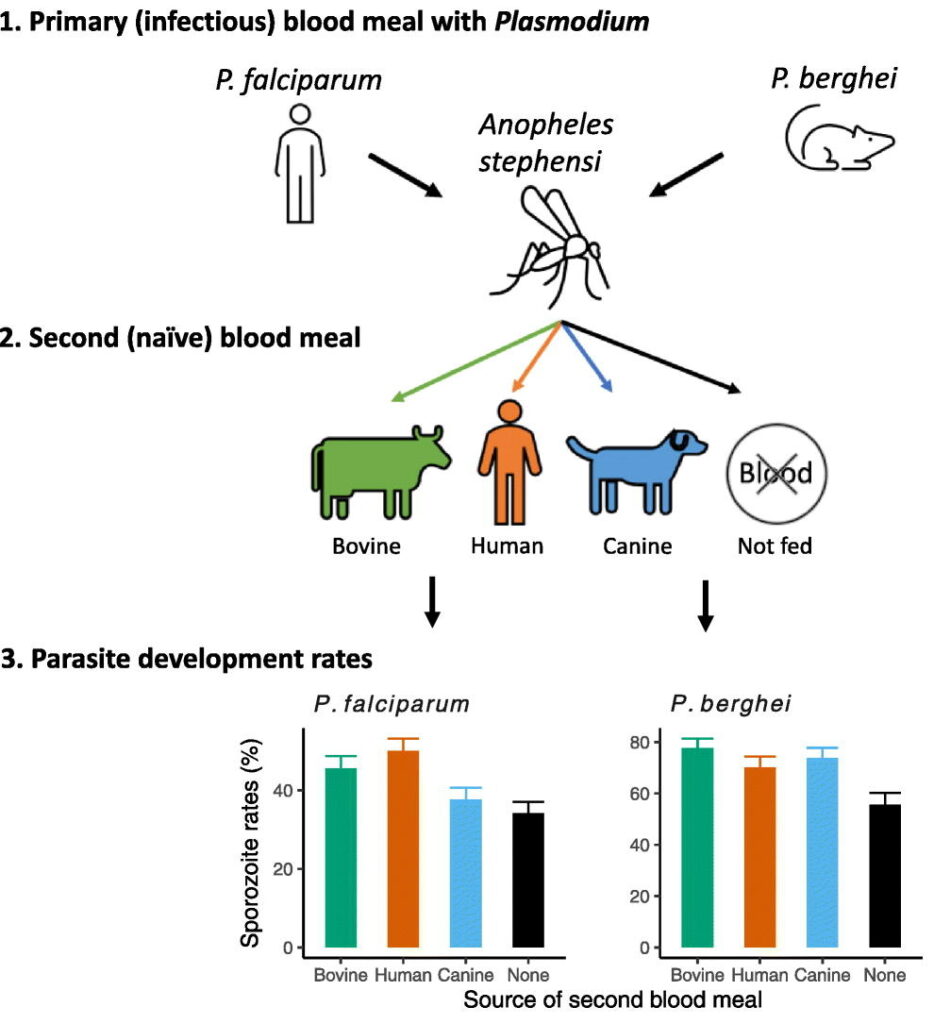
Testing Plasmodium vivax antimicrobial sensitivity is limited to ex vivo schizont maturation assays, which preclude determining the IC50s of delayed action antimalarials such as doxycycline. Using Plasmodium cynomolgi as a model for P. vivax, we determined the physiologically significant delayed death effect induced by doxycycline [IC50(96 h), 1,401 ± 607 nM]. As expected, IC50(96 h) to chloroquine (20.4 nM), piperaquine (12.6 µM), and tafenoquine (1,424 nM) were not affected by extended exposure.
Peter Christensen, Rosy Cinzah, Rossarin Suwanarusk, Adeline Chiew Yen Chua, Osamu Kaneko, Dennis E Kyle, Htin Lin Aung, Jessica Matheson, Pablo Bifani, Laurent Rénia, Gregory M Cook, Georges Snounou, Bruce Russell. Antimicrob Agents Chemother. 2024 Apr 8:e0028024. doi: 10.1128/aac.00280-24.