Trainee Adds New Tool to the Trypanosome Toolbox
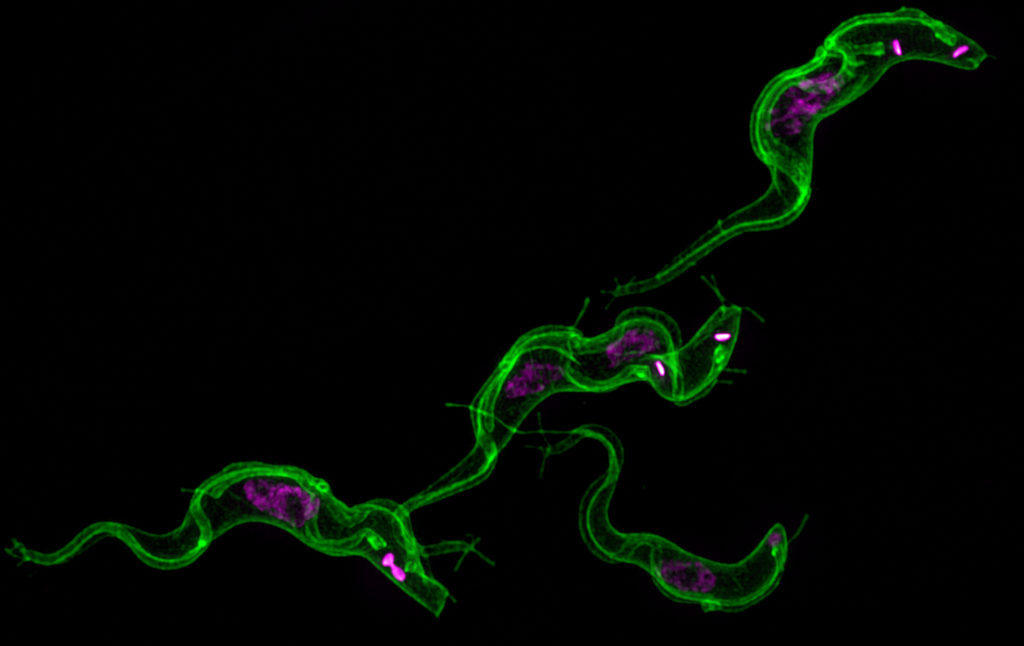
When Ph.D. trainee Justin Wiedeman started investigating the role of protein kinase TbCK1.2, an enzyme found near the flagellum of Trypanosoma brucei, he quickly ran into a problem common to parasitologists. He needed a better tool for visualizing the membranes of this parasite. Since none of the membrane probes on the market quite did the job, he looked at how he could modify one for his purpose. He found a successful candidate in Synaptic Systems’ mCLING.
What is Trypanosoma brucei?
Trypanosoma brucei is a single cell parasite that causes Human African Trypanosomiasis (HAT), which is also known as African sleeping sickness. HAT occurs in 36 sub-Saharan countries where tsetse flies transmit the parasite to people and livestock. In cattle, the disease is known as nagana. Tsetse fly control efforts have drastically reduced the number of cases. According to the World Health Organization, in 2015, there were around 2,800 cases. However, a person can be infected for months or even years without symptoms. By the time symptoms become evident, the person is in the advanced stages of the disease and their central nervous system is impaired.
New tools are needed to study trypanosomes
There is still much to be learned about the parasite that could lead to better detection and more effective treatment. A major obstacle to the study of this tiny organism is the lack of tools and technology. Kojo Mensa-Wilmot’s research group in the Center for Tropical and Emerging Global Diseases at The University of Georgia has been instrumental in developing techniques and tools to increase the research community’s understanding of T. brucei. Now, Wiedeman has added a new tool to the trypanosome biology toolbox – a general method of outlining trypanosomes in fluorescence microscopy experiments.
“We are the first group to solve this general problem in super-resolution microscopy of T. brucei,” said Wiedeman. “mCLING is a highly versatile tool for studying trypanosome biology – it can be used with live or fixed trypanosomes.”
Fluorescent microscopy has been a leading method of studying T. brucei; however, there are limitations to this technology. Super-resolution microscopy offers great advantages over standard fluorescence microscopy. By employing several techniques to increase resolution, it allows for the observation of objects smaller than what can be seen with visible light. Yet, it is not without its own limitations, most notably the inability to determine the periphery of cells. Without knowing the outer edges of the parasite, orientation of organelles and other structures within the cell is difficult.
“For Trypanosoma brucei, most of the membrane probes available do not work well in fixed trypanosomes,” said Wiedeman. “Researchers have been forced to use crude methods to outline trypanosomes in fluorescence microscopy.”
These “crude methods” include superimposing a transmitted light image or hand-drawing the outline. However, this workaround only allows for a two-dimensional study of the cell. Therefore, Wiedeman turned to a dye called mCLING that has been developed to track the membranes of neurons using super-resolution microscopy to see if he could adapt the technology to T. brucei membranes.
mCLING allows for the visualization of T. brucei membranes
“mCLING labels the flagellum and plasma membrane vividly, sometimes providing details of cell structure that rivals images obtained with scanning electron microscopy,” said Wiedeman.
Using a combination of standard-resolution and super-resolution fluorescence microscopy, he was able to confirm mCLING labels the plasma and flagellar membranes of T. brucei. Furthermore, using the Zeiss ELYRA S1 super-resolution microscopy in the Biomedical Microscopy Core, mCLING allowed for a 3D reconstruction of the parasite. This is the first time such an image has been reported. Finally, using the new ImageStream X Mark II in the CTEGD Cytometry Shared Resource Laboratory, he discovered mCLING could be used to track endocytosis (the process of importing molecules into the cell) in real time.
Recognizing mCLING’s potential to inform other studies of trypanosome biology, Weideman optimized protocols for using it with immunofluorescence assays and thus making possible what had been impossible with the overlay technique – visualizing the location of organelles in the vertical dimension relative to the cell body.
“It is especially well-suited for studying flagellar membrane biogenesis as well as kinetically tracking uptake of the plasma membrane into vesicles inside trypanosomes,” said Wiedeman. Other laboratories have already implemented these protocols in their own research. Steve Hajduk’s group, also in the Center for Tropical and Emerging Global Diseases, is using mCLING to study nanotubes in T. brucei.
This tool will allow for the study of trypanosomes in finer detail than ever before and the Mensa-Wilmot Research Group anticipates unlocking previously unseen secrets in T. brucei.
The full published study is available online: Wiedeman J, Mensa-Wilmot K (2018). A fixable probe for visualizing flagella and plasma membranes of the African trypanosome. PLoS One 13(5):e0197541. https://doi.org/10.1371/journal.pone.0197541
