Jessica Kissinger never set out to make databases. From the time she was a little girl, she wanted to be a biologist.
Today, the University of Georgia professor not only studies deadly pathogens like malaria and Cryptosporidium (a waterborne parasite), but also is a driving force behind worldwide, groundbreaking collaborations on novel databases. During her time at UGA, she has received nearly $40 million in federal and private grants and contracts.
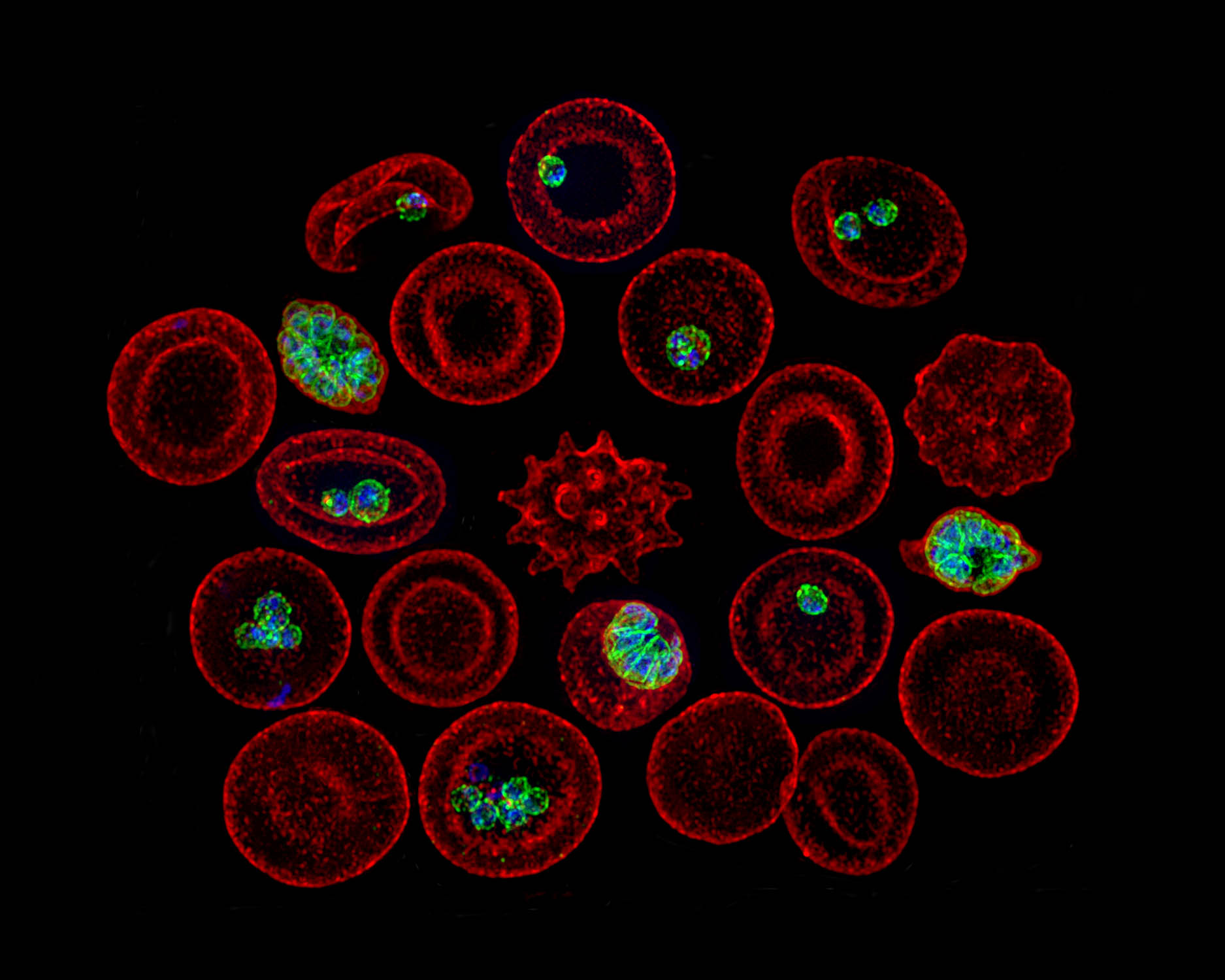
These databases can crunch vast amounts of biological information at warpspeed and reveal important patterns that pave the way for new approaches to scourges such as Leishmania (common in the tropics, subtropics, and southern Europe), toxoplasmosis (a systemic disease due to one of the world’s most common parasites), and Valley Fever (a fungus born on the wind that can cause lung and systemic infections). Novel drug and vaccine targets can be developed, as well as fresh insights on life-threatening pathogens.
“Fighting infections and developing new drug and vaccine targets requires detailed knowledge of a pathogen and how it functions,” explained Kissinger, a Distinguished Research Professor in UGA’s Department of Genetics, Institute of Bioinformatics and Center for Tropical and Emerging Global Diseases.
And, like internet searches, the databases are all free. Kissinger said it’s likely that pharmaceutical companies are mining some of the information in their quest to discover new therapeutic targets.
“They don’t tell us what they’re working on,” she said. “A database itself doesn’t produce a cure. A database can, however, remove most barriers to analysis of existing data.”
Big Data paves the way for big advances in science
It once took an entire decade to sequence a single genome—and the cost was many millions. Today, researchers can sequence a genome in a single afternoon for a few thousand dollars, transforming the field of genomics. Similar astounding advances have reshaped other ‘omics’ specialties, such as proteomics (study of proteins), metabolomics (study of metabolism), transcriptomics (study of RNA), and epigenomics (the influence of the environment on gene function). These advances mark the “Big Data” era in biology.
“The power that is unleashed by big data is phenomenal,” said Kissinger, “and it’s a very exciting time in history, with major funders and visionaries all across the world forming consortia to create a kind of ideal data universe.” Like explorers trekking into a new world, they will make discoveries we might only imagine right now.
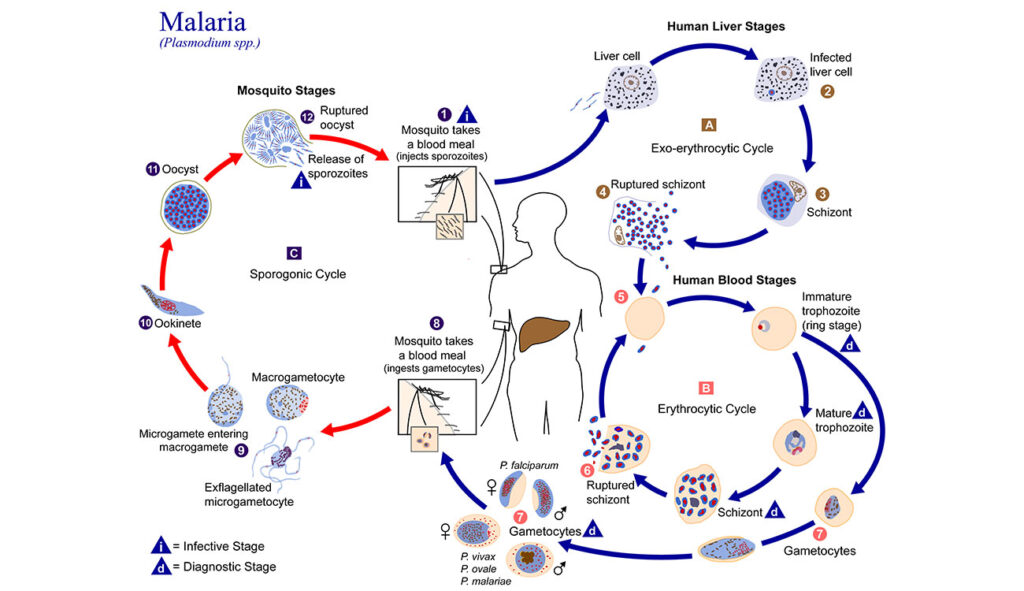
Creating a malaria database
Kissinger’s innovations began over 23 years ago, while she was a postdoctoral researcher at the University of Pennsylvania studying a single-celled parasite called Toxoplasma gondii. The parasite shares some important features with the malaria pathogen, whose genome was in the process of being sequenced.
“I rounded up genome data from all over the world on Plasmodium (the causative agent of malaria), and ran analyses and put it on a website, so I could study the genes it might share with Toxoplasma,” she recalled. “It turns out nobody had made the Plasmodium data available for searching before.”
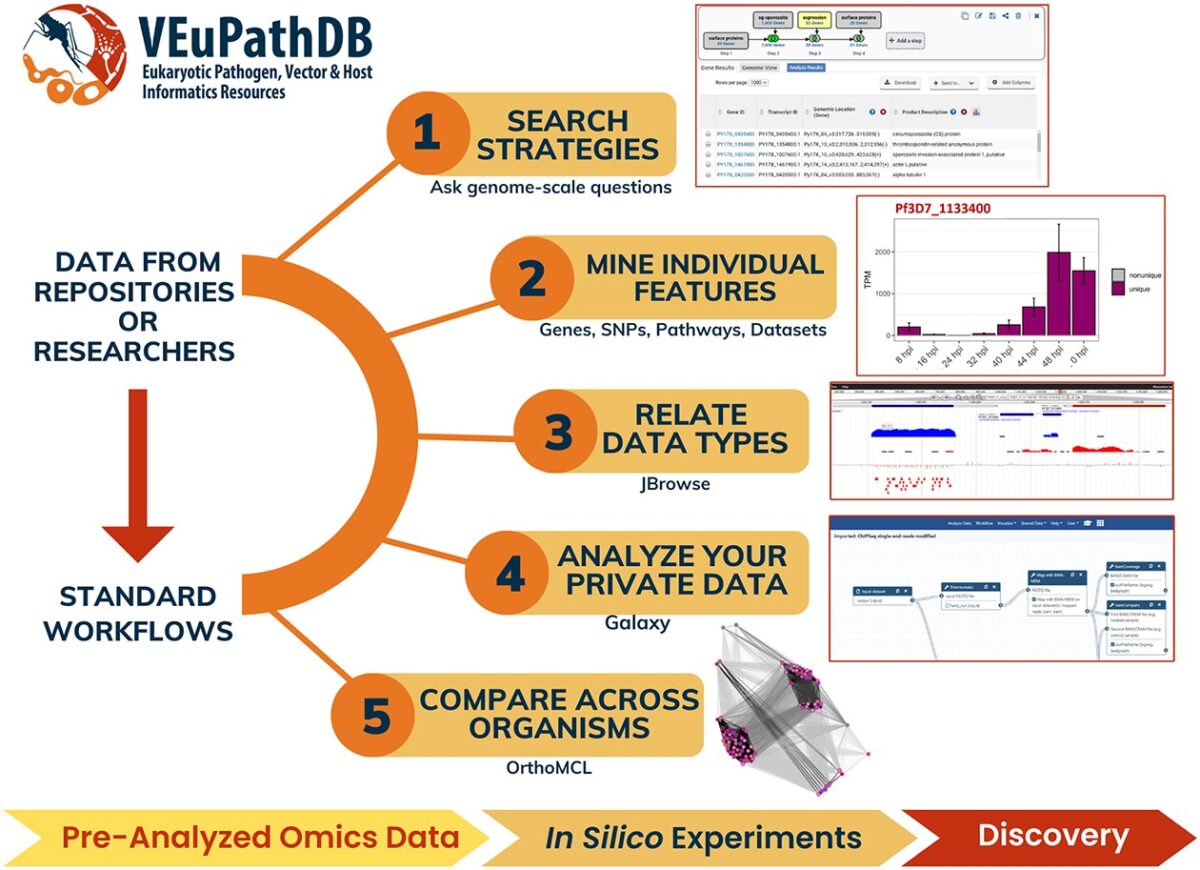
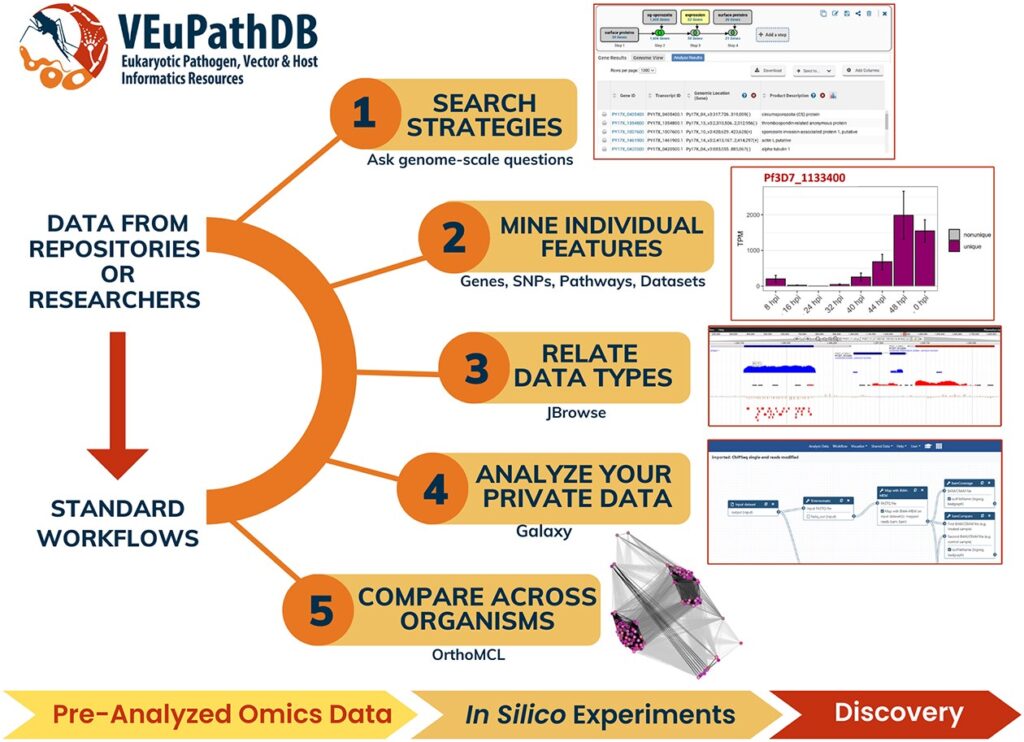
Soon she and her adviser, David Roos, had a million-dollar grant to formally establish a malaria database, PlasmoDB, and since its launch in 1999 it has grown to include additional pathogens and received continual funding from the NIH, the most recent for up to $38.4 million to maintain what has now become the Eukaryotic Pathogen, Vector and Host Informatics Resources knowledgebase (VEuPathDB), covering 14 different pathogens as well as host responses to infections. This comprehensive database is an integrated centralized resource for data mining on over 500 organisms.
The databases collectively contain over nine terabytes (9,000 gigabytes) of data, and have been compared to a Wikipedia for molecular parasitology by the British Society for Parasitology, which noted back in 2006: “We don’t know what we would do without it!”
Each month, VEuPathDB receives over 11 million hits from an average of 36,000 unique visitors in more than 100 countries, including India, Brazil and Kenya. A related database on vectors of disease (such as ticks that carry Lyme disease) was recently merged into VEuPathDB. The merger expanded each resource and enables researchers to better explore data on vectors such as ticks and mosquitoes and the pathogens they transmit.
Powerful tools are key to analyzing data
The databases are not just strings of numbers or words. They allow visualizations and graphic interfaces. Already, research is emerging that can help direct vaccine and drug development away from proteins that hosts and pathogens share, in order to protect the cell. Scientists using the databases have discovered proteins that reduce severe malaria and other proteins that protect malaria parasites from the human fever response. They have also found proteins that help Toxoplasma penetrate host cells.
In a single year an average of 200 publications a month cite VEuPathDB, and to date there have already been 24,000 citations total. Next up: cloud-ready applications and improved integration with yet other databases. These databases “have become essential data mining and access platforms for fungal and parasite genomics research,” said microbiologist and plant pathologist Jason Stajich of the University of California at Riverside.
“Without powerful, user-friendly tools to analyze it, “Big Data” is more a curse than a blessing,” explained John Boothroyd, an immunologist and microbiologist at Stanford University School of Medicine. “VEuPathDB is just such a tool and we owe Jessica Kissinger and her colleagues an enormous thank you for their tireless and selfless efforts to first conceive and then continuously improve this absolutely essential resource.”
Grants for related projects have come from a wide array of organizations, among them the Bill & Melinda Gates Foundation, the Sloan Foundation, and the World Health Organization. One of those projects, called ClinEpiDB, is home to a multicenter study that contains data from over 22,000 children from seven different sites in South Asia and Africa. This study is the largest ever to investigate the causes of diarrhea in children in lower- to middle-income countries. Other uses of ClinEpiDB include new data on hidden signs of malaria transmission in areas where incidence is declining, or how breastfeeding protects infants from common infections.
The VEuPathDB database would be enough to secure Kissinger’s reputation in the biological sciences, but she has not stopped there. At the University of Georgia, she was a founding member of the Institute of Bioinformatics, and served as its director from 2011 to 2109. The Institute’s mission is to facilitate cutting-edge interdisciplinary research in computational biology, and the program offers both masters and doctorates. She is a key researcher helping to partner a national hub for infectious disease research by linking with Emory University in Atlanta. The two institutions have grants totaling over $45 million to work on everything from tuberculosis to HIV to malaria.
“These databases are a success beyond my wildest dreams,” said Kissinger. “They are made by biologists for other biologists and address a real-life need.”