Database offers tool for global health collaborations
As the big data revolution continues to evolve, access to data that cut across many disciplines becomes increasingly valuable. In the field of public health, one barrier to sharing data is the need for users to fully comprehend complex methodological details and data variables in order to properly conduct analyses.
The Clinical Epidemiology Database, ClinEpiDB.org, aims to address these barriers by not only providing access to huge volumes of data, but also providing tools to help interpret complex global epidemiologic research studies. The development of ClinEpiDB has been led by the University of Georgia’s Institute of Bioinformatics, University of Pennsylvania’s School of Arts and Sciences and its Perelman School of Medicine, and the University of Liverpool’s Institute of Integrative Biology.
On March 7, ClinEpiDB released data, methodology and documentation from “The Etiology, Risk Factors, and Interactions of Enteric Infections and Malnutrition and the Consequences for Child Health and Development” (MAL-ED) study. The MAL-ED study represents a nearly decade-long research collaboration between the Foundation for the National Institutes of Health (FNIH), Fogarty International Center, and an international network of investigators.
The MAL-ED study was designed to help identify environmental exposures early in a child’s life that are associated with shortfalls in physical growth, cognitive development, and immunity. The study characterizes gut function biomarkers on the causal pathway from environmental exposure to growth and development deficits and assesses diversity across geographic locations with respect to exposures and child health and development. The MAL-ED consortium has published a significant library of peer-reviewed publications and ClinEpiDB now makes the MAL-ED data highly visible and accessible in new and exciting ways.
“It is great to see how investments and effort directed at data being Findable, Accessible, Interoperable and Reusable—i.e., F.A.I.R—are beginning to bear fruit,” said Jessica Kissinger, UGA Distinguished Research Professor of Genetics and co-principal investigator on the Bill & Melinda Gates Foundation award that funded the ClinEPi Development. “Too many important studies are buried in the scientific or medical literature and not easily accessible or reusable in moving the frontier in the important battles related to infectious disease and human health. This multi-institutional, multiple-funder, interdisciplinary approach is working.”
ClinEpiDB is also home to the Global Enteric Multicenter Study (GEMS) which contains data from more than 22,000 children from seven sites in South Asia and Africa and was the largest-ever study to investigate the causes to moderate-to-severe diarrheal illness in children in lower- to middle-income countries. The most recent ClinEpiDB release also contains data from GEMS1A, a continuation of the GEMS study that broadened its scope to include less-severe diarrheal episodes. The addition of MAL-ED adds to the growing resource of high-quality maternal and child global health data.
“Over 10 years, our international network of investigators collaborated through MAL-ED to better understand the complicated relationships among intestinal infections, nutrition and other environmental exposures on child development,” said Michael Gottlieb, FNIH deputy director of science (retired) and lead PI for the MAL-ED study. “The MAL-ED Network generated a high-quality data set, possibly the largest of its kind, on various research areas from cognitive abilities to gut function to immunological response. We are pleased to make this dataset available through ClinEpiDB so it can be used by researchers far into the future to increase scientific understanding, test new research hypotheses and design and implement better intervention strategies to reduce childhood morbidity and mortality.”
MAL-ED sites (located in Iquitos, Peru; Fortaleza, Brazil; Haydom, Tanzania; Limpopo, South Africa; Bhaktapur, Nepal; Naushero Feroze, Pakistan; Vellore, India; Dhaka, Bangladesh) allowed for comparisons to be made among and between children living in geographically and culturally diverse urban and rural environments and in countries at different levels of economic development.
MAL-ED data in ClinEpiDB account for over 1.3 million observations covering anthropometrics, nutrition, vaccination status, diarrheal and respiratory disease episodes and countless other details collected by community field workers in 2009-2014. The current release includes longitudinal data from children followed two times a week for the first 24 months of life.
Future data releases will contain data for some children up to 5 years of age. ClinEpiDB allows users to walk through these data easily via an intuitive interface, enabling point-and-click filtering, simple queries and more complex “search strategies.”
See https://youtu.be/535PcFrBH8M for a video introduction to this resource. ClinEpiDB will continue to grow and provide increased access to malaria and maternal and child health global datasets thus facilitating epidemiologic research in an open data environment while protecting patient identity.
Data boost: Using big data to fight disease

From leisure to health, digital databases can streamline nearly every facet of modern life.
Remember when making travel plans to a single destination took hours? Now booking flights, hotels and rental cars is just a few clicks—and a credit card—away thanks to travel sites like Expedia, Travelocity and others. Travelers get to compare competitors on price, amenities, customer reviews and proximity to popular locations. The sites pull together multiple data points from various sources (such as pricing from the seller, reviews from users, and maps from Google) and organize them for customers to view.
Jessica Kissinger, the director of UGA’s Institute for Bioinformatics and member of the Center for Tropical and Emerging Global Diseases, is doing for infectious disease research what travel sites did for vacation planning.
All over the world, researchers are racing to stop the spread of deadly and debilitating pathogens such as malaria. As those researchers and public health officials determine, or record data about a disease, Kissinger and her colleagues work to make that data accessible and searchable by the global research community for free.
“We take data generated by others and make them better,” says Kissinger, a Distinguished Research Professor of Genetics. More specifically, Kissinger and a team of cell biologists, geneticists and computer scientists pull disease data from a variety of sources, translate them into standard formats and make them searchable.
Jessica Kissinger, the director of UGA’s Institute for Bioinformatics, is doing for infectious disease research what travel sites did for vacation planning.
Enabling Discovery
How does building a database fight disease? Data help researchers construct and test their ideas about how to create treatments for diseases or map out ways to halt their spread.
“We don’t give them answers,” Kissinger says. “We give them a framework in which to generate and test hypotheses.”
Kissinger and her team have built databases to take on malaria and other infectious diseases such as toxoplasmosis, cryptosporidiosis and trypanosomiasis. They are also creating tools for studying childhood malnutrition and factors related to disease, and making them accessible to all as they become publicly available. These databases collectively service more than 70,000 unique users a month from more than 100 countries.
To put it simply, her work saves time. It speeds the pace discovery for the next possible solution, the next cure. Without these databases, researchers could spend weeks, months, even years researching existing literature on a disease in the library or recreating work in the lab.

These are tools by biologist for biologists … I think it is that sense of being a member of that community, having your finger on the pulse of what’s going on, that allows you to keep the tools useful. ~ Jessie Kissinger Director, Institute of Bioinformatics
Career Evolution
Kissinger didn’t set out to build databases.
She was trained as a molecular evolutionary biologist, not a computer scientist. “I like to see how molecules change over time,” she says. “When I started in school it was about how a gene or protein evolved.”
It turned out that her field was evolving too. Technology was allowing researchers to understand molecules through bigger data sets. Now, scientists aren’t just looking at individual genes but entire genomes, which are the complete sets of genes in a cell or organism.
As the field evolved, Kissinger learned and embraced the technology. Over time, she shifted her balance away from the so called “wet lab,” where she worked directly with the organisms, to focus mostly on the computer-based “dry lab.”
Her database work started with malaria and continued to expand.
“Now we make 10 different component databases for over 300 organisms (EuPathDB.org), a comparative database to see how conserved genes are across organisms and a new epidemiology database to study the prevalence, spread and factors related to disease in humans (ClinEpiDB.org).” she says.
Kissinger’s team relies on an expert advisory board that helps the researchers customize the databases for each disease community, so they have the largest impact on research. It helps that Kissinger started in a wet lab before diving into informatics.
“These are tools by biologist for biologists,” she says. “We have a lot of computer scientists in the middle, but I think it is that sense of being a member of that community, having your finger on the pulse of what’s going on, that allows you to keep the tools useful.”
SUPPORT OUR RESEARCH
Give to the Center Tropical & Emerging Global Diseases General Fund
[button size=’large’ style=” text=’Give Now’ icon=” icon_color=” link=’https://ctegd.uga.edu/give/’ target=’_self’ color=” hover_color=” border_color=” hover_border_color=” background_color=’#b80d32′ hover_background_color=” font_style=” font_weight=” text_align=’center’ margin=”]
Originally published at https://greatcommitments.uga.edu/story/data-boost/

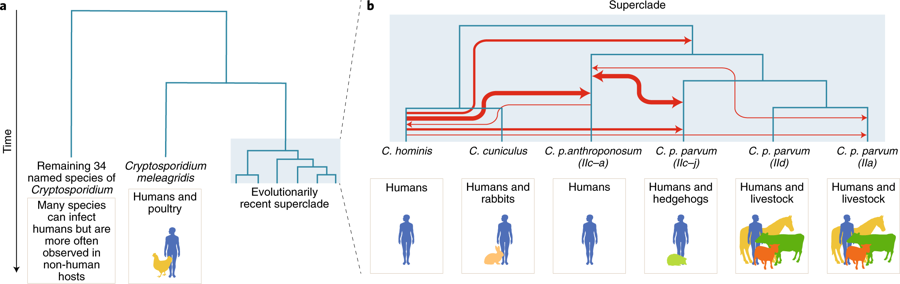 A large-scale comparative genomic survey of Cryptosporidium species and subtypes reveals a cryptic anthroponotic Cryptosporidium parvum branch and a large, recent superclade of species and subtypes that undergo genetic exchange, potentially facilitating host associations.
A large-scale comparative genomic survey of Cryptosporidium species and subtypes reveals a cryptic anthroponotic Cryptosporidium parvum branch and a large, recent superclade of species and subtypes that undergo genetic exchange, potentially facilitating host associations.