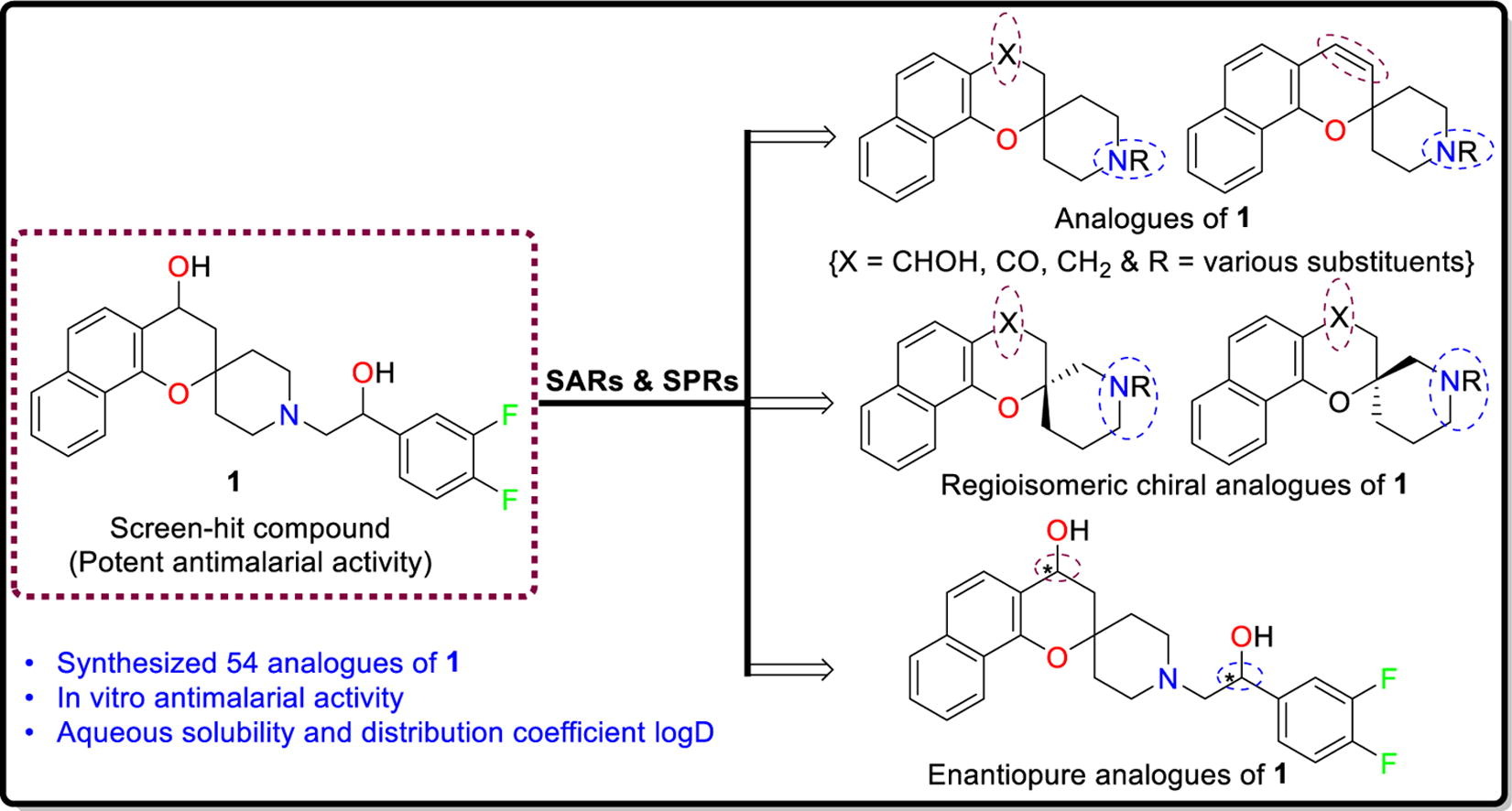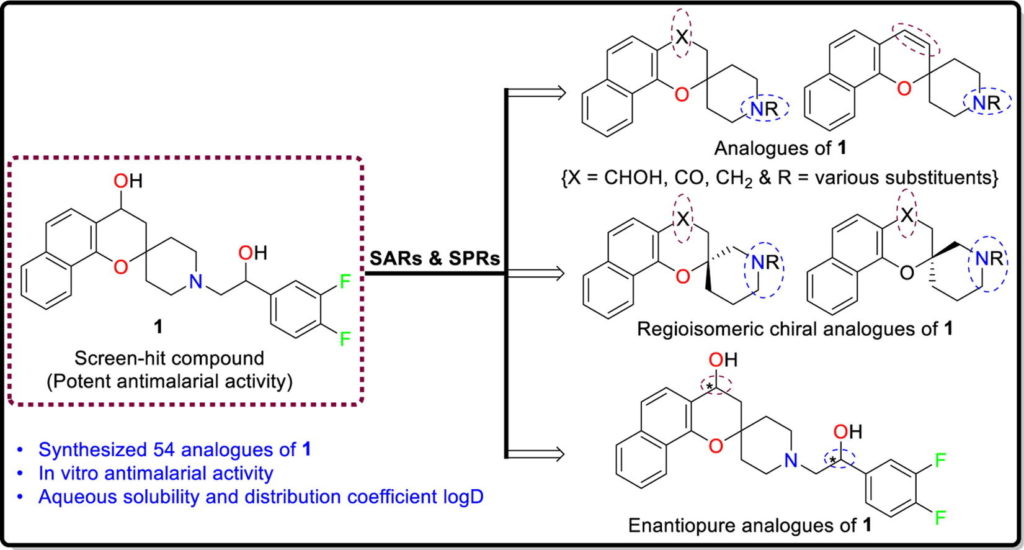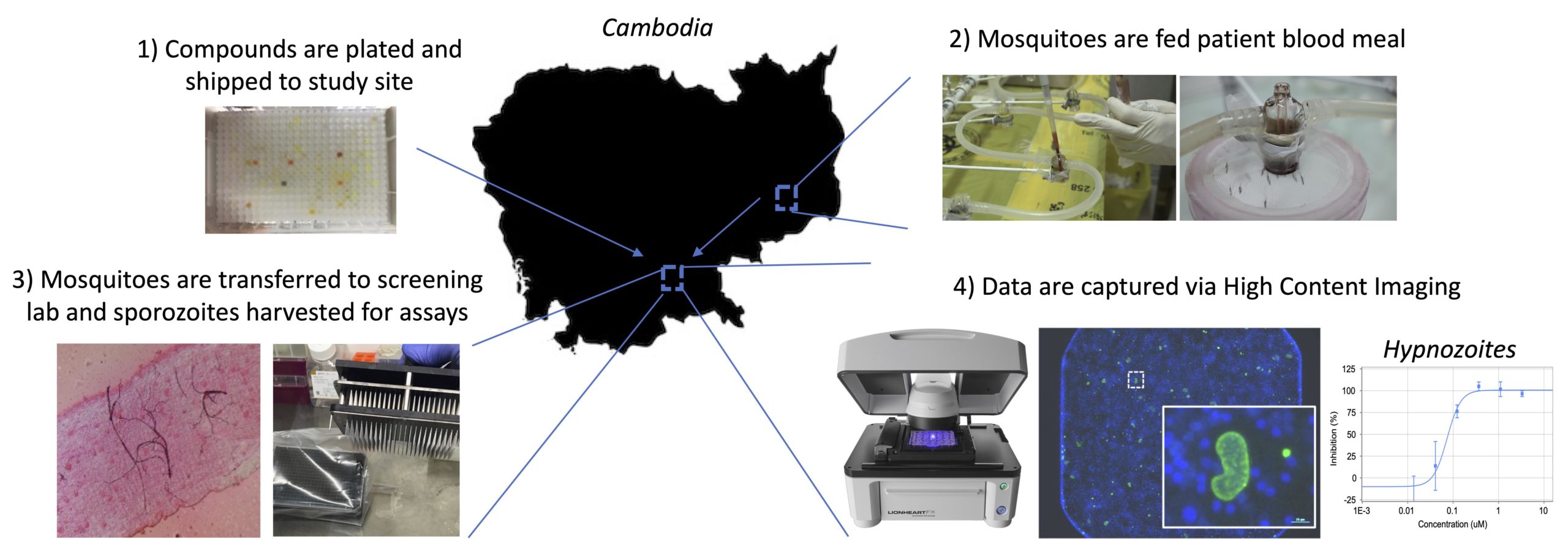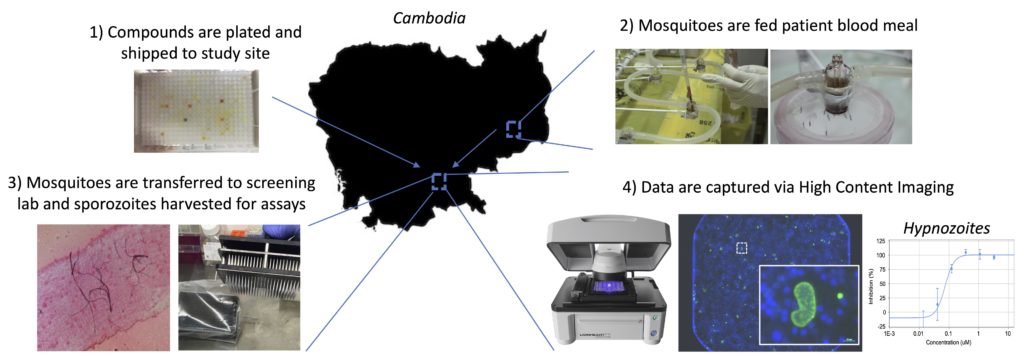
Justine Shiau, an NIH T32 fellow in Dr. Dennis Kyle’s laboratory, is originally from Taipei, Taiwan, and moved to the states after elementary school. She received her bachelor’s degree in Biology from the Pennsylvania State University, where she became interested in disease transmission, disease ecology, and parasitology while working with Dr. Ashutosh Pathak. Upon graduation, she moved to Athens to continue her training with Dr. Pathak, who at that time was working in the transmission ecology of vector-borne diseases with Dr. Courtney Murdock. Over the next two years, she took part in research projects revolving around vector biology and mosquito-transmitted pathogens. She was accepted by the UGA Integrated Life Science graduate program in Fall 2018.
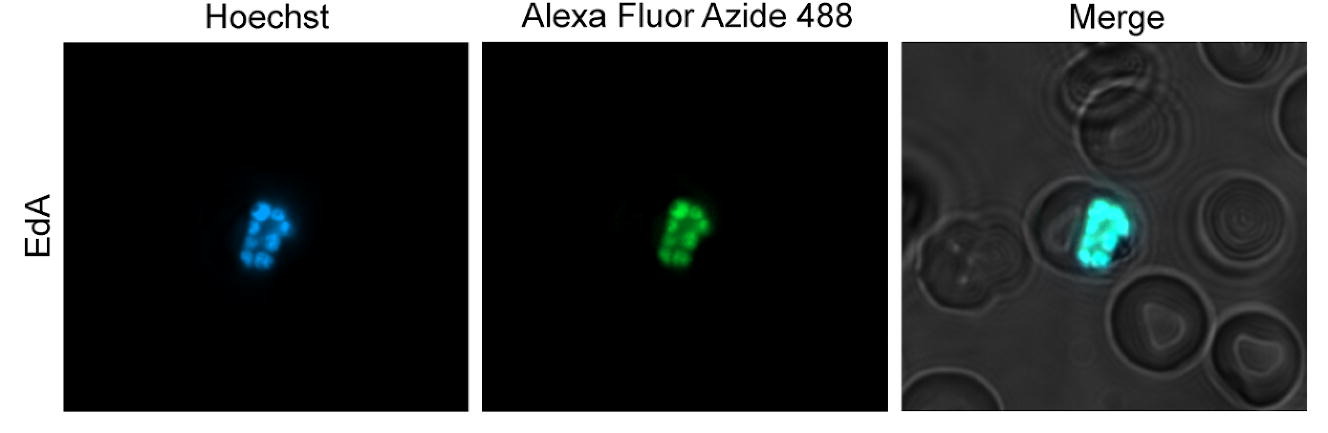
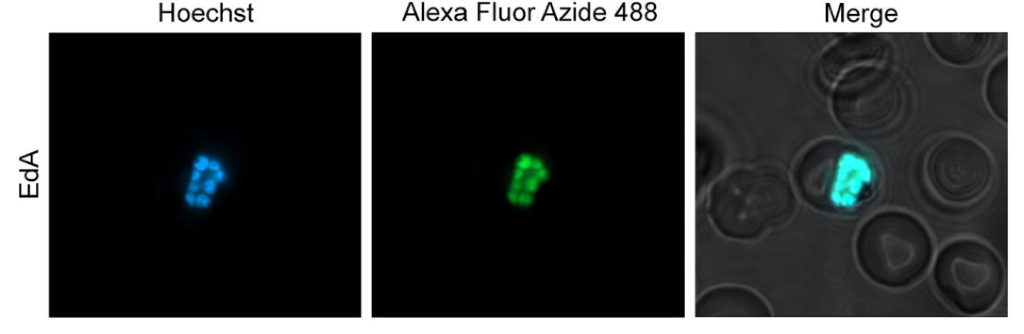
In the Kyle lab, Justine is currently working on the transmission stages of Plasmodium falciparum, a human malaria parasite that causes significant mortality worldwide, specifically on the biology of the parasite transitioning from the vector to the human and the early stages within the human, prior to disease onset. She aims to complete the parasite’s life cycle in a laboratory setting, which would be a powerful tool to help further our understanding of the host-parasite interactions. She hopes to better understand the parasite biology and the transmission dynamic that the mosquitoes could have on the downstream infection in humans, which can potentially help us better understand and combat this horrible disease.
Why did you choose UGA?
UGA has one of the finest insectary facilities that allows the transmission of Plasmodium falciparum. Additionally, the Center of Tropical and Emerging Global Diseases (CTEGD) is the hub for parasitologists. The Center provides state-of-the-art infrastructure, research equipment, and, most of all, a supportive environment to cultivate and train graduate students to meet our goals.
What is your research focus?
Plasmodium falciparum is a parasite that causes malaria, which 50% of the world’s population is at risk of getting. Many children die from malaria every year; we cannot effectively prevent diseases and transmissions without a well-rounded understanding of the parasite’s biology and the essential players (mosquitoes) to complete its life cycle. My overarching goal is to complete the parasite’s life cycle in the lab. Currently, we are focusing on the biology of the parasite and its transition from mosquito back to human and within the human: from liver-to-blood stage infections. While doing this, there are two primary objectives that I would like to meet. First, I want to better understand the important factors for the parasites to establish infection in the human liver cells. Second, I am curious whether the mosquito stage infection can also impact the parasite’s efficiency in establishing infection in the human liver.
What are your future professional plans?
After graduate school, I hope to continue my postdoctoral training. I would like to pursue interdisciplinary research, with crosstalk between disease-ecology, parasitology, and vector biology.
Any advice for a student interested in this field?
Be open-minded and respectful to people with different expertise and people with diverse backgrounds.
Support trainees like Justine by giving today to the Center for Tropical & Emerging Global Diseases.