Plastid Biogenesis in Malaria Parasites Requires the Interactions and Catalytic Activity of the Clp Proteolytic System
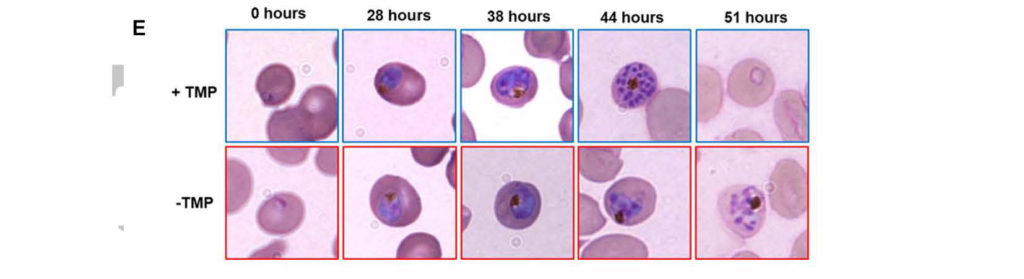
The human malaria parasite, Plasmodium falciparum, contains an essential plastid called the apicoplast. Most apicoplast proteins are encoded by the nuclear genome and it is unclear how the plastid proteome is regulated. Here, we study an apicoplast-localized caseinolytic-protease (Clp) system and how it regulates organelle proteostasis. Using null and conditional mutants, we demonstrate that the P. falciparum Clp protease (PfClpP) has robust enzymatic activity that is essential for apicoplast biogenesis. We developed a CRISPR/Cas9-based system to express catalytically dead PfClpP, which showed that PfClpP oligomerizes as a zymogen and is matured via transautocatalysis. The expression of both wild-type and mutant Clp chaperone (PfClpC) variants revealed a functional chaperone-protease interaction. Conditional mutants of the substrate-adaptor (PfClpS) demonstrated its essential function in plastid biogenesis. A combination of multiple affinity purification screens identified the Clp complex composition as well as putative Clp substrates. This comprehensive study reveals the molecular composition and interactions influencing the proteolytic function of the apicoplast Clp system and demonstrates its central role in the biogenesis of the plastid in malaria parasites.
Anat Florentin, Dylon R. Stephens, Carrie F. Brooks, Rodrigo P. Baptista, and Vasant Muralidharan. Proc Natl Acad Sci USA. 2020 Jun 1;201919501. doi: 10.1073/pnas.1919501117.
Directing Traffic: Chaperone-mediated protein transport in malaria parasites
The ability of eukaryotic parasites from the phylum Apicomplexa to cause devastating diseases is predicated upon their ability to maintain faithful and precise protein trafficking mechanisms. Their parasitic life cycle depends on the trafficking of effector proteins to the infected host cell, transport of proteins to several critical organelles required for survival, as well as transport of parasite and host proteins to the digestive organelles to generate the building blocks for parasite growth. Several recent studies have shed light on the molecular mechanisms parasites utilize to transform the infected host cells, transport proteins to essential metabolic organelles, and for biogenesis of organelles required for continuation of their life cycle. Here, we review key pathways of protein transport originating and branching from the endoplasmic reticulum, focusing on the essential roles of chaperones in these processes. Further, we highlight key gaps in our knowledge that prevents us from building a holistic view of protein trafficking in these deadly human pathogens.
Anat Florentin, David W. Cobb, Heather M. Kudyba, Vasant Muralidharan. Cell Microbiol. 2020 May 9:e13215. doi: 10.1111/cmi.13215.
An Endoplasmic Reticulum CREC Family Protein Regulates the Egress Proteolytic Cascade in Malaria Parasites
The endoplasmic reticulum (ER) is thought to play an essential role during egress of malaria parasites because the ER is assumed to be required for biogenesis and secretion of egress-related organelles. However, no proteins localized to the parasite ER have been shown to play a role in egress of malaria parasites. In this study, we generated conditional mutants of the Plasmodium falciparum endoplasmic reticulum-resident calcium-binding protein (PfERC), a member of the CREC family. Knockdown of the PfERC gene showed that this gene is essential for asexual growth of P. falciparum Analysis of the intraerythrocytic life cycle revealed that PfERC is essential for parasite egress but is not required for protein trafficking or calcium storage. We found that PfERC knockdown prevents the rupture of the parasitophorous vacuole membrane. This is because PfERC knockdown inhibited the proteolytic maturation of the subtilisin-like serine protease SUB1. Using double mutant parasites, we showed that PfERC is required for the proteolytic maturation of the essential aspartic protease plasmepsin X, which is required for SUB1 cleavage. Further, we showed that processing of substrates downstream of the proteolytic cascade is inhibited by PfERC knockdown. Thus, these data establish that the ER-resident CREC family protein PfERC is a key early regulator of the egress proteolytic cascade of malaria parasites.
IMPORTANCE The divergent eukaryotic parasites that cause malaria grow and divide within a vacuole inside a host cell, which they have to break open once they finish cell division. The egress of daughter parasites requires the activation of a proteolytic cascade, and a subtilisin-like protease initiates a proteolytic cascade to break down the membranes blocking egress. It is assumed that the parasite endoplasmic reticulum plays a role in this process, but the proteins in this organelle required for egress remain unknown. We have identified an early ER-resident regulator essential for the maturation of the recently discovered aspartic protease in the egress proteolytic cascade, plasmepsin X, which is required for maturation of the subtilisin-like protease. Conditional loss of PfERC results in the formation of immature and inactive egress proteases that are unable to breakdown the vacuolar membrane barring release of daughter parasites.
Manuel A. Fierro, Beejan Asady, Carrie F. Brooks, David W. Cobb, Alejandra Villegas, Silvia N. J. Moreno, Vasant Muralidharan. mBio. 2020 Feb 25;11(1). pii: e03078-19. doi: 10.1128/mBio.03078-19.
The ER chaperone PfGRP170 is essential for asexual development and is linked to stress response in malaria parasites
 The vast majority of malaria mortality is attributed to one parasite species: Plasmodium falciparum. Asexual replication of the parasite within the red blood cell is responsible for the pathology of the disease. In Plasmodium, the endoplasmic reticulum (ER) is a central hub for protein folding and trafficking as well as stress response pathways. In this study, we tested the role of an uncharacterized ER protein, PfGRP170, in regulating these key functions by generating conditional mutants. Our data show that PfGRP170 localizes to the ER and is essential for asexual growth, specifically required for proper development of schizonts. PfGRP170 is essential for surviving heat shock, suggesting a critical role in cellular stress response. The data demonstrate that PfGRP170 interacts with the Plasmodium orthologue of the ER chaperone, BiP. Finally, we found that loss of PfGRP170 function leads to the activation of the Plasmodium eIF2α kinase, PK4, suggesting a specific role for this protein in this parasite stress response pathway.
The vast majority of malaria mortality is attributed to one parasite species: Plasmodium falciparum. Asexual replication of the parasite within the red blood cell is responsible for the pathology of the disease. In Plasmodium, the endoplasmic reticulum (ER) is a central hub for protein folding and trafficking as well as stress response pathways. In this study, we tested the role of an uncharacterized ER protein, PfGRP170, in regulating these key functions by generating conditional mutants. Our data show that PfGRP170 localizes to the ER and is essential for asexual growth, specifically required for proper development of schizonts. PfGRP170 is essential for surviving heat shock, suggesting a critical role in cellular stress response. The data demonstrate that PfGRP170 interacts with the Plasmodium orthologue of the ER chaperone, BiP. Finally, we found that loss of PfGRP170 function leads to the activation of the Plasmodium eIF2α kinase, PK4, suggesting a specific role for this protein in this parasite stress response pathway.
Heather M. Kudyba, David W. Cobb, Manuel A. Fierro, Anat Florentin, Dragan Ljolje, Balwan Singh, Naomi W. Lucchi, Vasant Muralidharan. 2019. Cell Microbiol.:e13042. doi: 10.1111/cmi.13042
Field evaluation of malaria malachite green loop-mediated isothermal amplification in health posts in Roraima state, Brazil
BACKGROUND:
Microscopic detection of malaria parasites is the standard method for clinical diagnosis of malaria in Brazil. However, malaria epidemiological surveillance studies specifically aimed at the detection of low-density infection and asymptomatic cases will require more sensitive and field-usable tools. The diagnostic accuracy of the colorimetric malachite green, loop-mediated, isothermal amplification (MG-LAMP) assay was evaluated in remote health posts in Roraima state, Brazil.
METHODS:
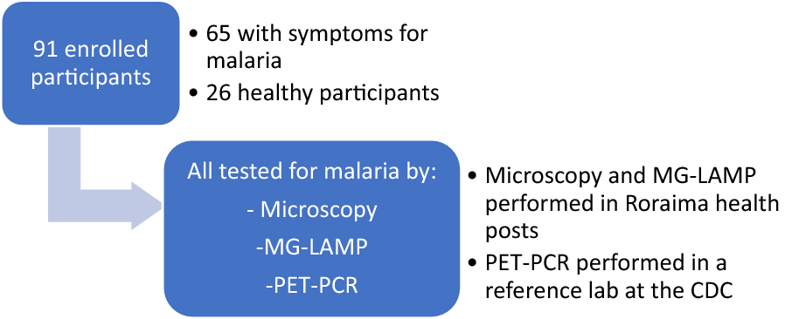
Study participants were prospectively enrolled from health posts (healthcare-seeking patients) and from nearby villages (healthy participants) in three different study sites. The MG-LAMP assay and microscopy were performed in the health posts. Two independent readers scored the MG-LAMP tests as positive (blue/green) or negative (clear). Sensitivity and specificity of local microscopy and MG-LAMP were calculated using results of PET-PCR as a reference.
RESULTS:
A total of 91 participants were enrolled. There was 100% agreement between the two MG-LAMP readers (Kappa = 1). The overall sensitivity and specificity of MG-LAMP were 90.0% (95% confidence interval (CI) 76.34-97.21%) and 94% (95% CI 83.76-98.77%), respectively. The sensitivity and specificity of local microscopy were 83% (95% CI 67.22-92.66%) and 100% (95% CI 93.02-100.00%), respectively. PET-PCR detected six mixed infections (infection with both Plasmodium falciparum and Plasmodium vivax); two of these were also detected by MG-LAMP and one by microscopy. Microscopy did not detect any Plasmodium infection in the 26 healthy participants; MG-LAMP detected Plasmodium in five of these and PET-PCR assay detected infection in three. Overall, performing the MG-LAMP in this setting did not present any particular challenges.
CONCLUSION:
MG-LAMP is a sensitive and specific assay that may be useful for the detection of malaria parasites in remote healthcare settings. These findings suggest that it is possible to implement simple molecular tests in facilities with limited resources.
Heather M. Kudyba, Jaime Louzada, Dragan Ljolje, Karl A. Kudyba, Vasant Muralidharan, Joseli Oliveira-Ferreira, and Naomi W. Lucchi. 2019. Malar J. 2019 Mar 25;18(1):98. doi: 10.1186/s12936-019-2722-1.
Plasmodium falciparum cGMP-dependent protein kinase interacts with a subunit of the parasite proteasome
ABSTRACT
Malaria is caused by the protozoan parasite Plasmodium, which undergoes a complex life cycle in a human host and a mosquito vector. The parasite’s cyclic GMP (cGMP)-dependent protein kinase (PKG) is essential at multiple steps of the life cycle. Phosphoproteomic studies in Plasmodium falciparum erythrocytic stages and Plasmodium berghei ookinetes have identified proteolysis as a major biological pathway dependent on PKG activity. To further understand PKG’s mechanism of action, we screened a yeast two-hybrid library for P. falciparum proteins that interact with P. falciparum PKG (PfPKG) and tested peptide libraries to identify its phosphorylation site preferences. Our data suggest that PfPKG has a distinct phosphorylation site and that PfPKG directly phosphorylates parasite RPT1, one of six AAA+ ATPases present in the 19S regulatory particle of the proteasome. PfPKG and RPT1 interact in vitro, and the interacting fragment of RPT1 carries a PfPKG consensus phosphorylation site; a peptide carrying this consensus site competes with the RPT1 fragment for binding to PfPKG and is efficiently phosphorylated by PfPKG. These data suggest that PfPKG’s phosphorylation of RPT1 could contribute to its regulation of parasite proteolysis. We demonstrate that proteolysis plays an important role in a biological process known to require Plasmodium PKG: invasion by sporozoites of hepatocytes. A small-molecule inhibitor of proteasomal activity blocks sporozoite invasion in an additive manner when combined with a Plasmodium PKG-specific inhibitor. Mining the previously described parasite PKG-dependent phosphoproteomes using the consensus phosphorylation motif identified additional proteins that are likely to be direct substrates of the enzyme.
K. Govindasamy, R. Khan, M. Snyder, H. J. Lou, P. Du, H. M. Kudyba, V. Muralidharan, B. E. Turk, P. Bhanot. 2018. Infection and Immunity. https://doi.org/10.1128/IAI.00523-18
CRISPR/Cas9 Gene Editing to Make Conditional Mutants of Human Malaria Parasite P. falciparum
ABSTRACT
Malaria is a significant cause of morbidity and mortality worldwide. This disease, which primarily affects those living in tropical and subtropical regions, is caused by infection with Plasmodium parasites. The development of more effective drugs to combat malaria can be accelerated by improving our understanding of the biology of this complex parasite. Genetic manipulation of these parasites is key to understanding their biology; however, historically the genome of P. falciparum has been difficult to manipulate. Recently, CRISPR/Cas9 genome editing has been utilized in malaria parasites, allowing for easier protein tagging, generation of conditional protein knockdowns, and deletion of genes. CRISPR/Cas9 genome editing has proven to be a powerful tool for advancing the field of malaria research. Here, we describe a CRISPR/Cas9 method for generating glmS-based conditional knockdown mutants in P. falciparum. This method is highly adaptable to other types of genetic manipulations, including protein tagging and gene knockouts.
Kudyba, H. M., Cobb, D. W., Florentin, A., Krakowiak, M., Muralidharan, V. 2018. J. Vis. Exp. (139), e57747, doi:10.3791/57747
Trainee Spotlight: Anat Florentin

Anat Florentin, a post-doctoral associate in Vasant Muralidharan‘s laboratory, is originally from Israel. She received her BSc degree from Tel-Aviv University and MSc from the Weizmann Institute of Science. She obtained her Ph.D. also from the Weizmann Institute where she studied programmed cell death mechanisms using the fruit fly as a model organism. Dr. Florentin moved to the United States 4 years ago when she joined the Muralidharan Research Group. During her time at UGA, she has received a number of awards in recognition of her research:
- American Heart Association Postdoctoral Fellowship (2018-2020)
- Postdoctoral Research Award, UGA Office of Research (2018)
- Foreign travel award, UGA Office of Research (2018)
- Best Poster Presentation award at the UGA GSPS Research Day (2016)
- Best Postdoctoral Poster award at the 2015 UGA Conference on Drug Discovery (2015)
Why did you choose UGA?
Since my background is in basic cell biology and genetics, I knew very little of the biology of parasites but was determined to study malaria. While I was looking into different places in Europe and the US, I met with another Israeli, Lilach Sheiner who, at the time, was doing her postdoctoral training with Dr. Boris Striepen at UGA. She told me very good things of CTEGD and of a great newly recruited faculty who studies malaria. I came for a visit, and was impressed by the engaging scientific community, the super friendly atmosphere and the variety of different parasites and approaches to study tropical neglected diseases. I am so glad I made this decision!
What is your research focus/project and why are you interested in the topic?
The goal of my research is to understand the unique cell biology of malaria parasites and to identify potential drug targets. In order to do that I develop and apply genetic and molecular tools that are used to manipulate the genome of the parasite. During my years in the lab I was involved in several projects; One of them studies mechanisms by which the parasite transports proteins into the host red blood cell. Another interesting project focuses on a conserved complex from bacterial origin that resides within a unique parasite organelle called the apicoplast. Lastly, I am looking for genes that might be involved in programmed cell death processes in the parasites.
What are your future professional plans?
I want to establish my own research lab, conduct independent research and train the next generation of future scientists.
Have you done any field work or is there a collaborator/field site that you would like to visit in order to enhance your training?
Although we use field samples in our studies, I have never been to any field site, and would absolutely love to visit one. I am positive it will enhance my training and will add another layer to the work that I am doing. I am sure that visiting any field site in a malaria endemic area, such as Africa or Southeast Asia would be an enriching experience that would underline the significance of our work.
What is your favorite thing about UGA and Athens, GA?
Many things… At CTEGD I cherish the collaborative atmosphere, the variety of parasitism-related topics, the strong basic science that goes together with field studies and translational research. I am highly appreciative of the fact that I have access to a huge amount of knowledge by working side by side with top experts in these fields.
Athens is also great. Moving here in 2014 with a family of 2 young kids couldn’t go smoother! We found here a great community of friends, great public schools, and amazing nature. I love the mountains, the trees and the wildlife around us!
Any advice for students interested in this field?
There is still so much to do and learn in the field of parasitology and every discovery that you make may impact the life of the millions that suffer from these diseases. Don’t hesitate if you don’t know much about parasites. No matter what your background is, you can use the tools and knowledge that you acquired and apply them to this challenging but rewarding research!
Your financial gift to the CTEGD Fund helps provide field research opportunities to trainees like Anat Florentin through The CTEGD Training Innovations in Parasitological Studies Fellowship.
[button size=’large’ style=” text=’Give Today’ icon=” icon_color=” link=’https://gail.uga.edu/commit?cat=campus&subcat=research&des=91700000′ target=’_self’ color=” hover_color=” border_color=” hover_border_color=” background_color=” hover_background_color=” font_style=” font_weight=” text_align=’center’ margin=”]
Anat Florentin receives 2018 Postdoctoral Research Award

Anat Florentin, a postdoctoral researcher in Vasant Muralidharan‘s laboratory at the Center for Tropical and Emerging Global Diseases, studies molecular mechanisms that drive life stages of Plasmodium falciparum, the deadliest of parasite species that infect humans with malaria. During her exceptionally productive years at UGA, she has advanced two related areas of research to learn more about the functions of P. falciparum gene and metabolic pathways. First, she established a highly efficient, markerless system to create mutants more rapidly using the powerful CRISPR-Cas9-based genetic editing tool. Her data from this project was published in the high-impact journal mSphere. Second, she used the CRISPR-Cas9 tool to understand P. falciparum’s unique plastid known as the apicoplast, which harbors essential metabolic pathways for the parasite’s growth and whose biological processes could be ideal parasite-specific drug targets. This work has been recognized by multiple invitations to present her work and a first author publication in Cell Reports.
Created in 2011, the Postdoctoral Research Award recognizes the remarkable contributions of postdoctoral research scholars to the UGA research enterprise. The UGA Research Foundation funds up to two awards a year to current scholars.

