Regulatory T cell memory: implications for malaria
Regulatory T cells (Tregs) can persist as memory cells (mTregs) in both infectious and non-infectious settings. However, their functional behavior, phenotypic stability, and suppressive properties upon antigen re-exposure remain poorly understood. Emerging evidence suggests that mTregs exhibit enhanced proliferation and suppressive capacity upon re-encountering the same antigen, a feature that may be critical in recurrent …
Type I interferons induce guanylate-binding proteins and lysosomal defense in hepatocytes to control malaria
Plasmodium parasites undergo development and replication within hepatocytes before infecting erythrocytes and initiating clinical malaria. Although type I interferons (IFNs) are known to hinder Plasmodium infection within the liver, the underlying mechanisms remain unclear. Here, we describe two IFN-I-driven hepatocyte antimicrobial programs controlling liver-stage malaria. First, oxidative defense by NADPH oxidases 2 and 4 triggers …
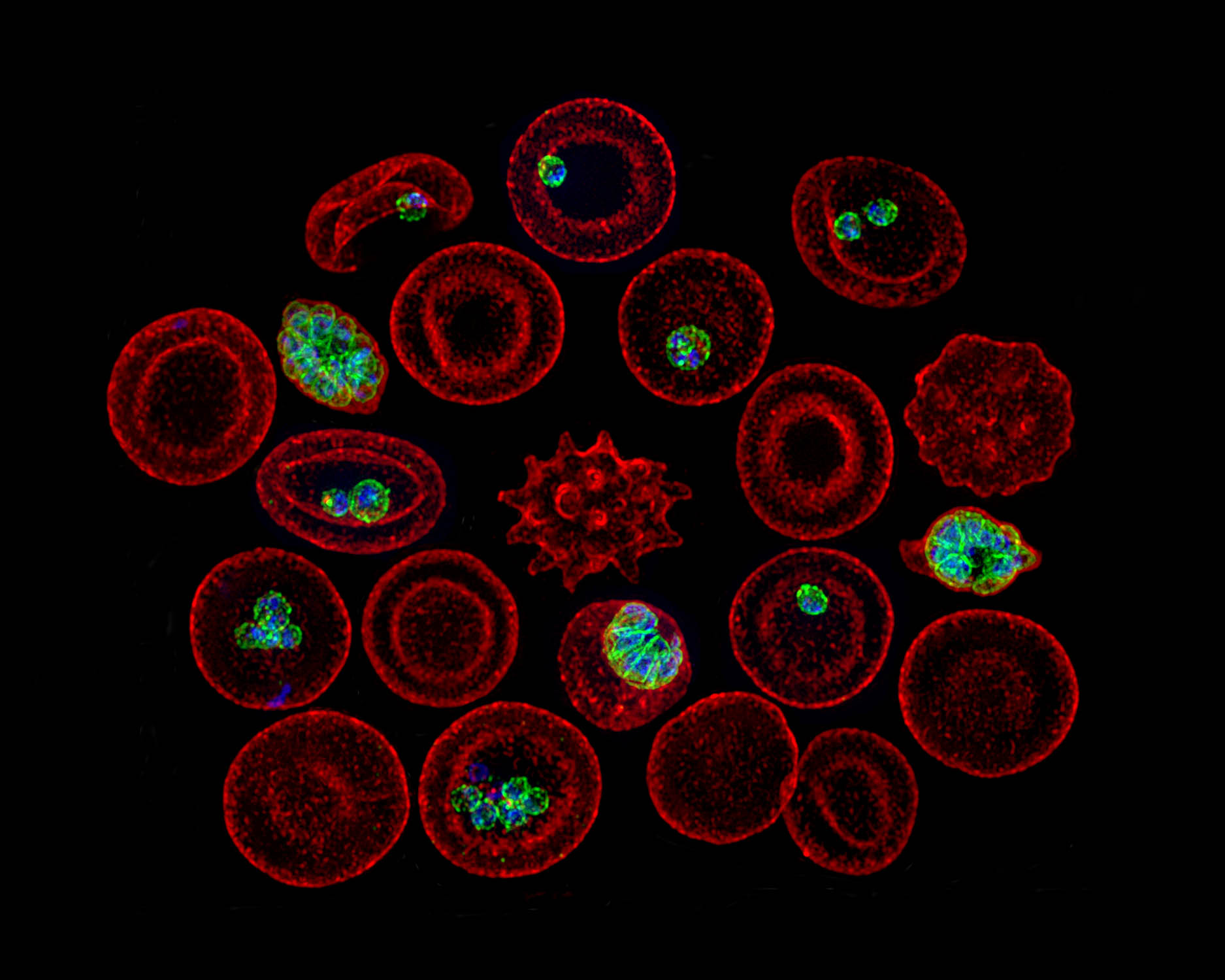
Hepatocytes and the art of killing Plasmodium softly
The Plasmodium parasites that cause malaria undergo asymptomatic development in the parenchymal cells of the liver, the hepatocytes, prior to infecting erythrocytes and causing clinical disease. Traditionally, hepatocytes have been perceived as passive bystanders that allow hepatotropic pathogens such as Plasmodium to develop relatively unchallenged. However, now there is emerging evidence suggesting that hepatocytes …
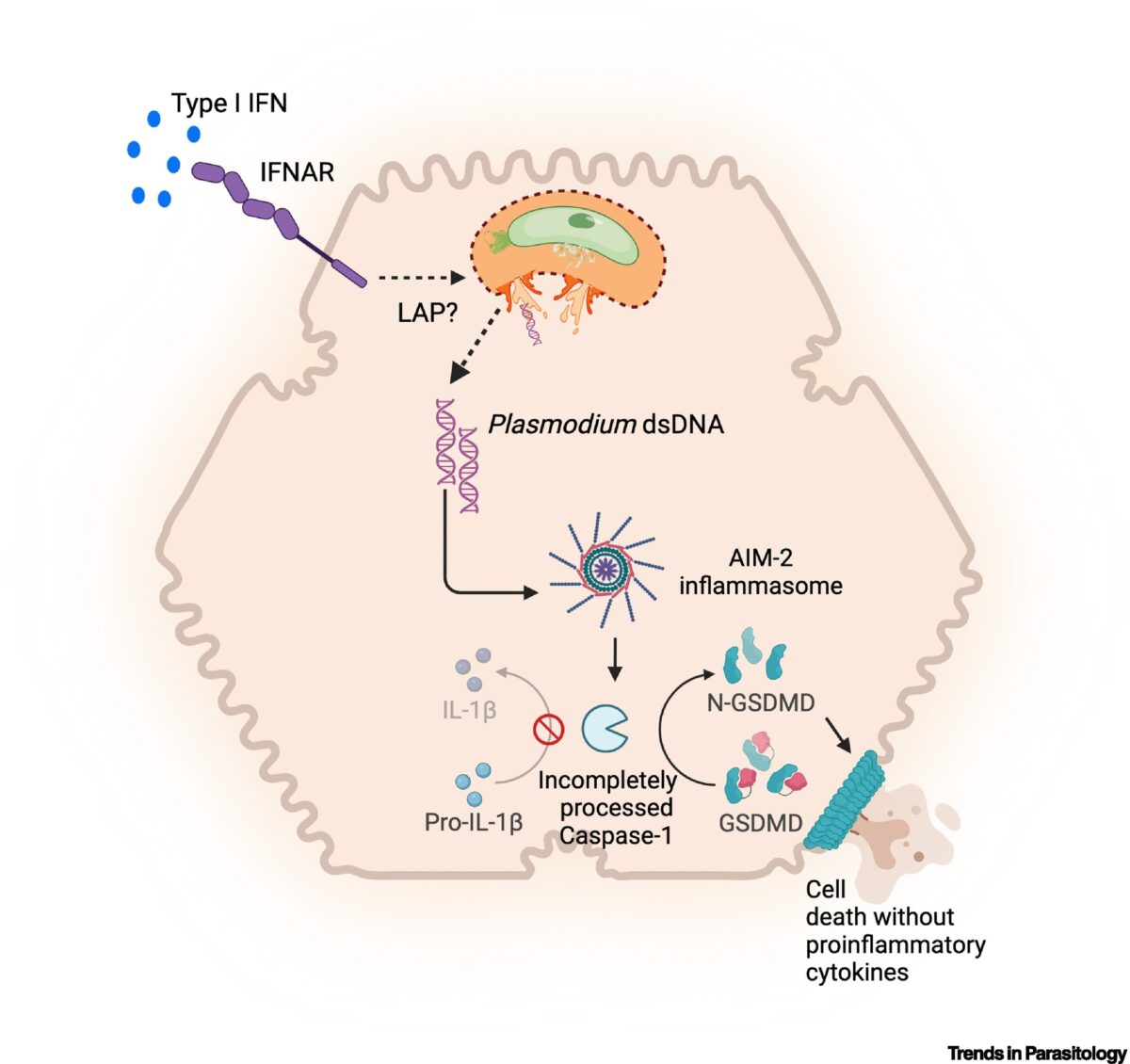
Inherently Reduced Expression of ASC Restricts Caspase-1 Processing in Hepatocytes and Promotes Plasmodium Infection
Inflammasome-mediated caspase-1 activation facilitates innate immune control of Plasmodium in the liver, thereby limiting the incidence and severity of clinical malaria. However, caspase-1 processing occurs incompletely in both mouse and human hepatocytes and precludes the generation of mature IL-1β or IL-18, unlike in other cells. Why this is so or how it impacts Plasmodium …
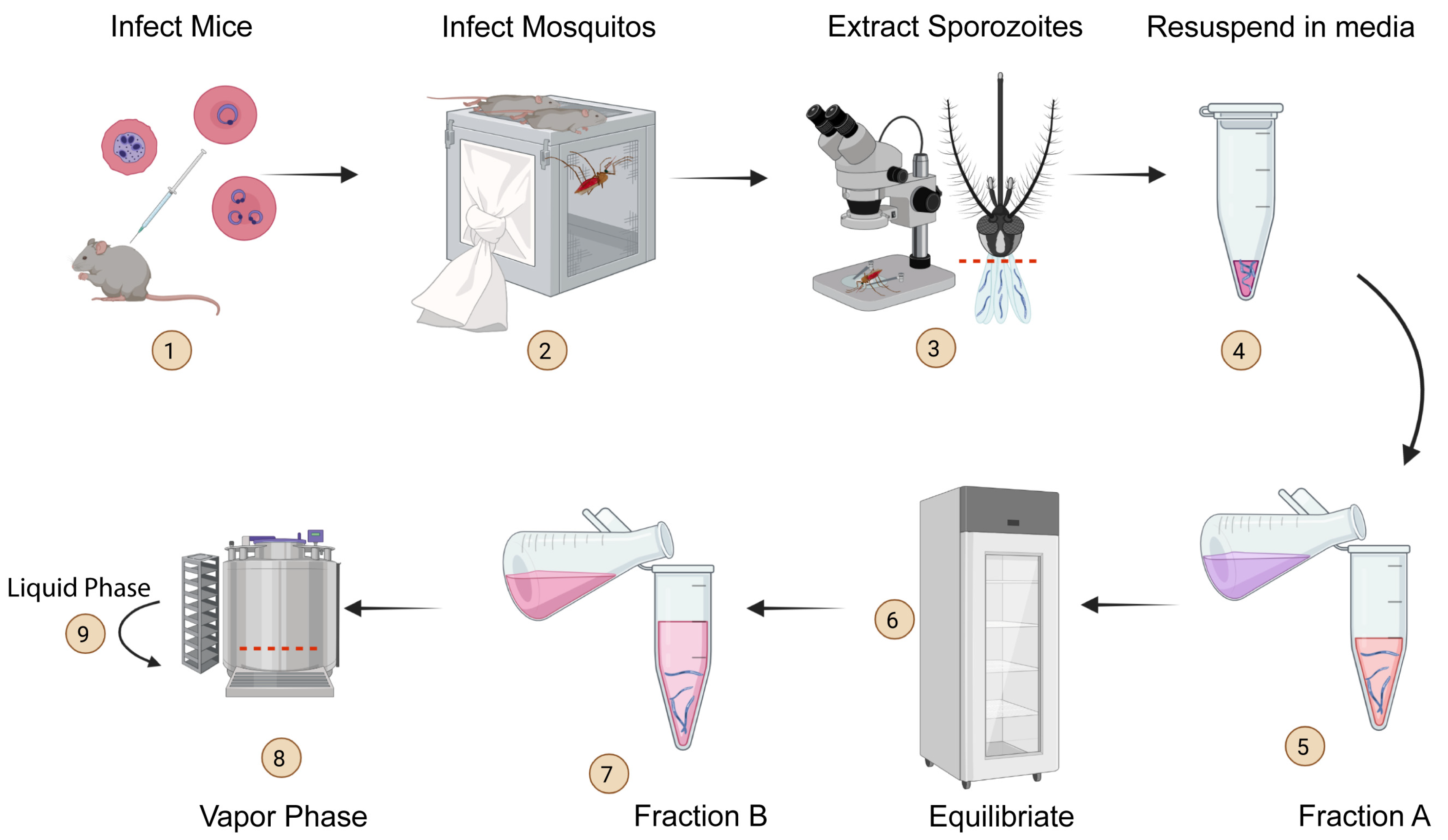
Generating Genetically Modified Plasmodium berghei Sporozoites
Malaria is a deadly disease caused by the parasite Plasmodium and is transmitted through the bite of female Anopheles mosquitoes. The sporozoite stage of Plasmodium deposited by mosquitoes in the skin of vertebrate hosts undergoes a phase of mandatory development in the liver before initiating clinical malaria. We know little about the biology of Plasmodium …