Ad libitum consumption of protein- or peptide-sucrose solutions stimulates egg formation by prolonging the vitellogenic phase of oogenesis in anautogenous mosquitoes
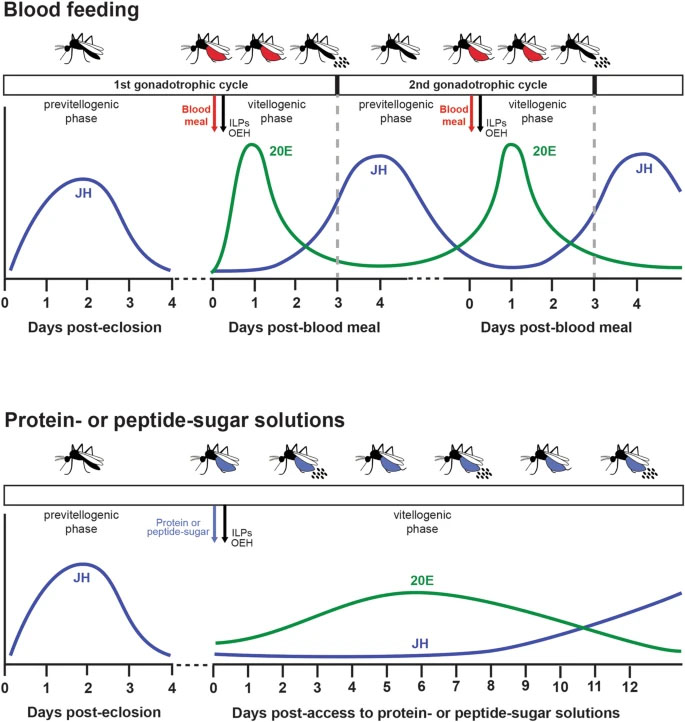 Background: Anautogenous mosquitoes commonly consume nectars and other solutions containing sugar but are thought to only produce eggs in discrete gonadotrophic cycles after blood-feeding on a vertebrate host. However, some anautogenous species are known to produce eggs if amino acids in the form of protein are added to a sugar solution. Unclear is how different sources of amino acids in sugar solutions affect the processes that regulate egg formation and whether responses vary among species. In this study, we addressed these questions by focusing on Aedes aegypti and conducting some comparative assays with Aedes albopictus, Anopheles gambiae, Anopheles stephensi and Culex quinquefasciatus.
Background: Anautogenous mosquitoes commonly consume nectars and other solutions containing sugar but are thought to only produce eggs in discrete gonadotrophic cycles after blood-feeding on a vertebrate host. However, some anautogenous species are known to produce eggs if amino acids in the form of protein are added to a sugar solution. Unclear is how different sources of amino acids in sugar solutions affect the processes that regulate egg formation and whether responses vary among species. In this study, we addressed these questions by focusing on Aedes aegypti and conducting some comparative assays with Aedes albopictus, Anopheles gambiae, Anopheles stephensi and Culex quinquefasciatus.
Methods: Adult female mosquitoes were fed sugar solutions containing amino acids, peptides or protein. Markers for activation of a gonadotrophic cycle including yolk deposition into oocytes, oviposition, ovary ecdysteroidogenesis, expression of juvenile hormone and 20-hydroxyecdysone-responsive genes, and adult blood-feeding behavior were then measured.
Results: The five anautogenous species we studied produced eggs when fed two proteins (bovine serum albumin, hemoglobin) or a mixture of peptides (tryptone) in 10% sucrose but deposited only small amounts of yolk into oocytes when fed amino acids in 10% sucrose. Focusing on Ae. aegypti, cultures were maintained for multiple generations by feeding adult females protein- or tryptone-sugar meals. Ad libitum access to protein- or tryptone-sugar solutions protracted production of ecdysteroids by the ovaries, vitellogenin by the fat body and protease activity by the midgut albeit at levels that were lower than in blood-fed females. Females also exhibited semi-continual oogenesis and repressed host-seeking behavior.
Conclusions: Several anautogenous mosquitoes produce eggs when provided ad libitum access to protein- or peptide-sugar meals, but several aspects of oogenesis also differ from females that blood-feed.
Ruby E Harrison, Kangkang Chen, Lilith South, Ange Lorenzi, Mark R Brown, Michael R Strand. Parasit Vectors. 2022 Apr 12;15(1):127. doi: 10.1186/s13071-022-05252-4.