Stable colonization of the model kissing bug Rhodnius prolixus by Trypanosoma cruzi Y strain
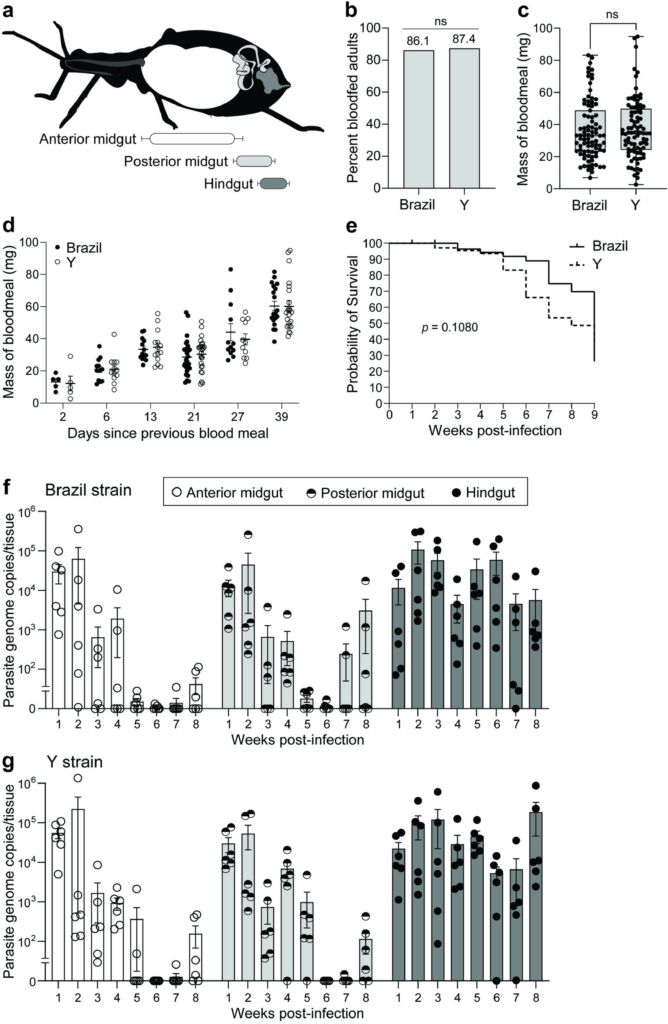
Trypanosoma cruzi is a single-celled eukaryotic parasite responsible for Chagas disease, a major cause of morbidity and mortality in Central and South America. While the host-pathogen interactions of T. cruzi have been extensively studied in vertebrate models, investigations into its interactions within its insect host remain limited. To address this gap and establish a genetically tractable system for studying parasite-vector dynamics, we conducted quantitative kinetic infection studies using the Y strain of T. cruzi and the model vector Rhodnius prolixus. We began by comparing parasite infection kinetics from two genetically diverse strains of T. cruzi, Brazil and Y, and demonstrated that ingested parasites from both strains transiently expand in the anterior regions of the insect digestive tract with stable colonization occurring in the hindgut over the long term. Notably, we demonstrated that the clonal Y strain, contrary to previous reports, can effectively infect and persist across multiple developmental stages of R. prolixus. Additionally, comparison of movement of parasites versus inert fluorescent microspheres introduced into artificial blood meals suggests that T. cruzi colonization of the R. prolixus gut occurs passively through peristaltic movement during digestion, rather than through active parasite-mediated chemotaxis. These findings highlight the T. cruzi Y strain – R. prolixus model system as a promising tool for the in-depth molecular characterization of parasite-vector interactions, potentially offering new insights into the biology of this neglected and deadly human pathogen.
Ruby E Harrison, Kevin J Vogel, Ronald Drew Etheridge. PLoS Negl Trop Dis. 2025 Mar 12;19(3):e0012906. doi: 10.1371/journal.pntd.0012906.