A new chromosome-level genome assembly and annotation of Cryptosporidium meleagridis

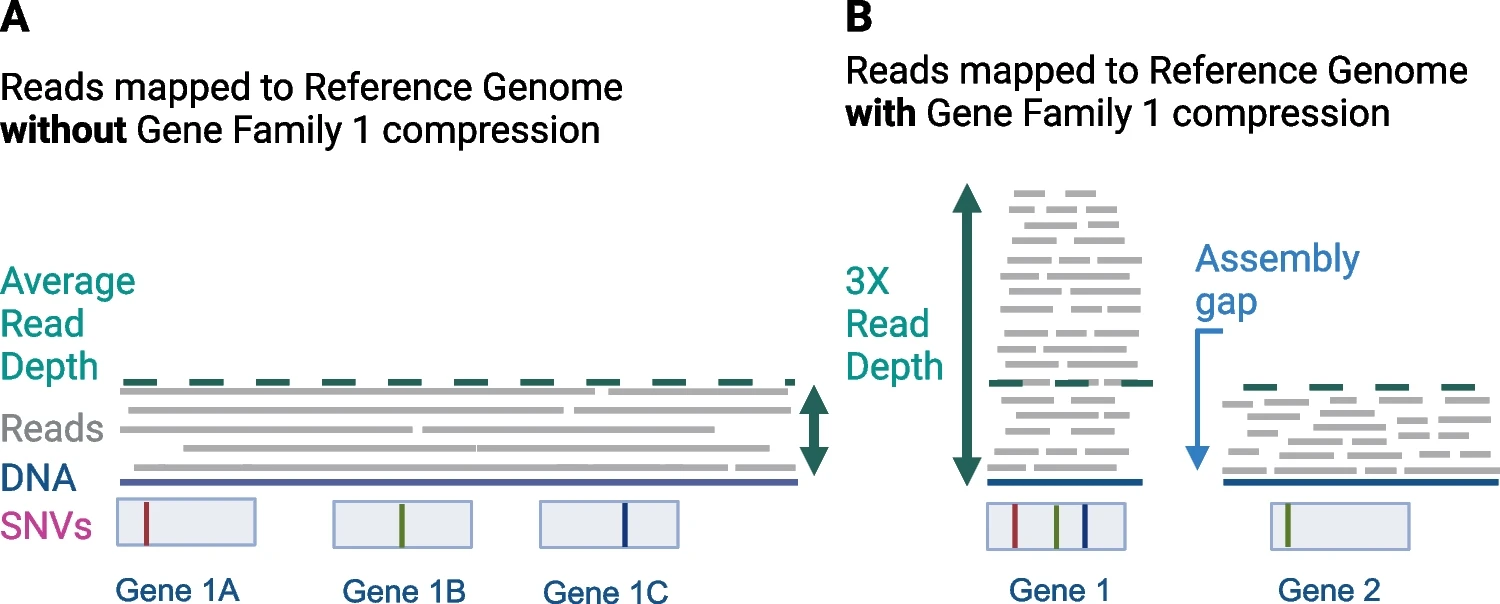
Cryptosporidium spp. are medically and scientifically relevant protozoan parasites that cause severe diarrheal illness in infants, immunosuppressed populations and many animals. Although most human Cryptosporidium infections are caused by C. parvum and C. hominis, there are several other human-infecting species including C. meleagridis, which are commonly observed in developing countries. Here, we annotated a hybrid long-read Oxford Nanopore Technologies and short-read Illumina genome assembly for C. meleagridis (CmTU1867) with DNA generated using multiple displacement amplification. The assembly was then compared to the previous C. meleagridis (CmUKMEL1) assembly and annotation and a recent telomere-to-telomere C. parvum genome assembly. The chromosome-level assembly is 9.2 Mb with a contig N50 of 1.1 Mb. Annotation revealed 3,919 protein-encoding genes. A BUSCO analysis indicates a completeness of 96.6%. The new annotation contains 166 additional protein-encoding genes and reveals high synteny to C. parvum IOWA II (CpBGF). The new C. meleagridis genome assembly is nearly gap-free and provides a valuable new resource for the Cryptosporidium community and future studies on evolution and host-specificity.
Lasya R Penumarthi, Rodrigo P Baptista, Megan S Beaudry, Travis C Glenn, Jessica C Kissinger. Sci Data. 2024 Dec 18;11(1):1388. doi: 10.1038/s41597-024-04235-7.