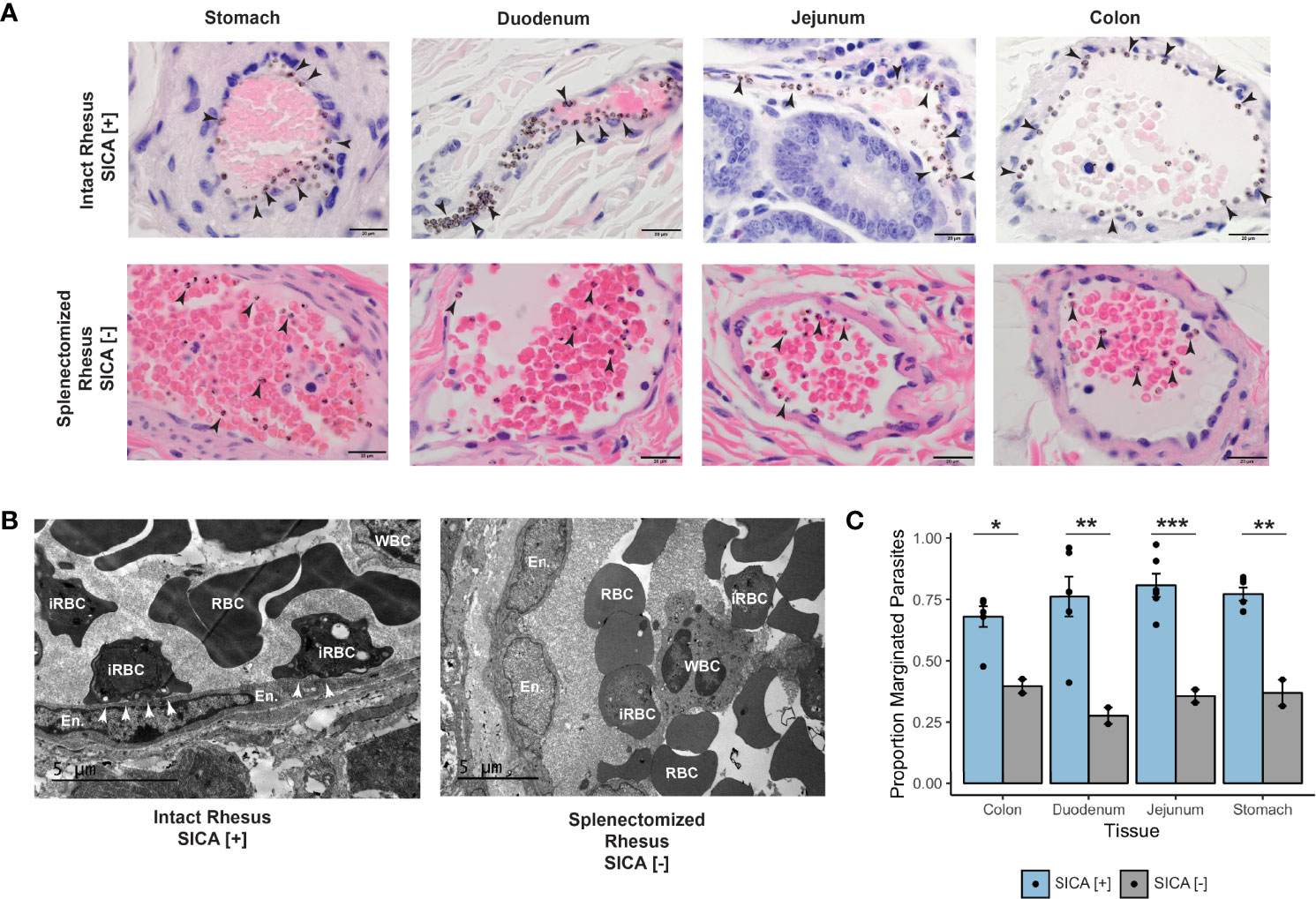
Malaria disrupts the rhesus macaque gut microbiome
Previous studies have suggested that a relationship exists between severity and transmissibility of malaria and variations in the gut microbiome, yet only limited information exists on the temporal dynamics of the gut microbial community during a malarial infection. Here, using a rhesus macaque model of relapsing malaria, we investigate how malaria affects the gut microbiome. …