Genome Editing by CRISPR/Cas9 in Trypanosoma cruzi
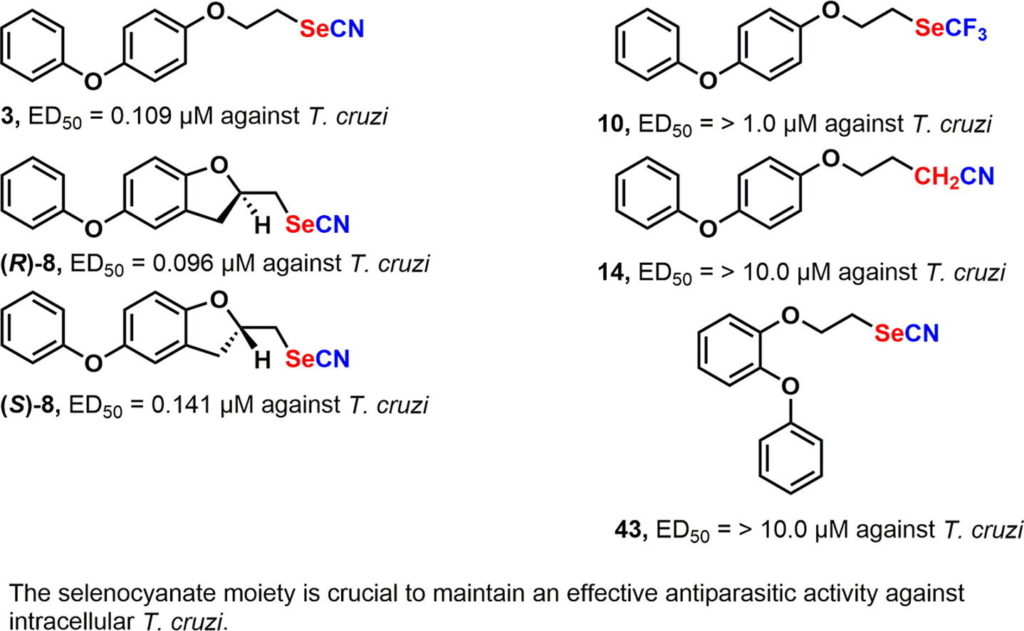
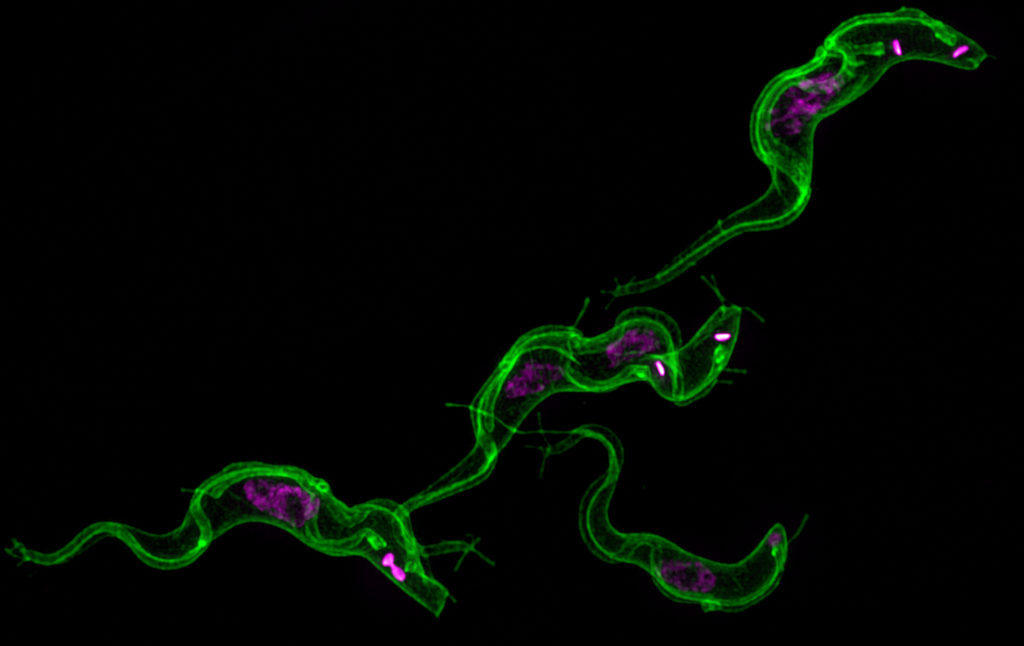
The genetic manipulation of the human parasite Trypanosoma cruzi has been significantly improved since the implementation of the CRISPR/Cas9 system for genome editing in this organism. The system was initially used for gene knockout in T. cruzi, later on for endogenous gene tagging and more recently for gene complementation. Mutant cell lines obtained by CRISPR/Cas9 have been used for the functional characterization of proteins in different stages of this parasite’s life cycle, including infective trypomastigotes and intracellular amastigotes. In this chapter we describe the methodology to achieve genome editing by CRISPR/Cas9 in T. cruzi. Our method involves the utilization of a template cassette (donor DNA) to promote double-strand break repair by homologous directed repair (HDR). In this way, we have generated homogeneous populations of genetically modified parasites in 4–5 weeks without the need of cell sorting, selection of clonal populations, or insertion of more than one resistance marker to modify both alleles of the gene. The methodology has been organized according to three main genetic purposes: gene knockout, gene complementation of knockout cell lines generated by CRISPR/Cas9, and C-terminal tagging of endogenous genes in T. cruzi. In addition, we refer to the specific results that have been published using each one of these strategies.
Noelia Lander, Miguel A. Chiurillo, Roberto Docampo. 2019. Methods Mol Biol. 2019;1955:61-76. doi: 10.1007/978-1-4939-9148-8_5