Data boost: Using big data to fight disease

From leisure to health, digital databases can streamline nearly every facet of modern life.
Remember when making travel plans to a single destination took hours? Now booking flights, hotels and rental cars is just a few clicks—and a credit card—away thanks to travel sites like Expedia, Travelocity and others. Travelers get to compare competitors on price, amenities, customer reviews and proximity to popular locations. The sites pull together multiple data points from various sources (such as pricing from the seller, reviews from users, and maps from Google) and organize them for customers to view.
Jessica Kissinger, the director of UGA’s Institute for Bioinformatics and member of the Center for Tropical and Emerging Global Diseases, is doing for infectious disease research what travel sites did for vacation planning.
All over the world, researchers are racing to stop the spread of deadly and debilitating pathogens such as malaria. As those researchers and public health officials determine, or record data about a disease, Kissinger and her colleagues work to make that data accessible and searchable by the global research community for free.
“We take data generated by others and make them better,” says Kissinger, a Distinguished Research Professor of Genetics. More specifically, Kissinger and a team of cell biologists, geneticists and computer scientists pull disease data from a variety of sources, translate them into standard formats and make them searchable.
Jessica Kissinger, the director of UGA’s Institute for Bioinformatics, is doing for infectious disease research what travel sites did for vacation planning.
Enabling Discovery
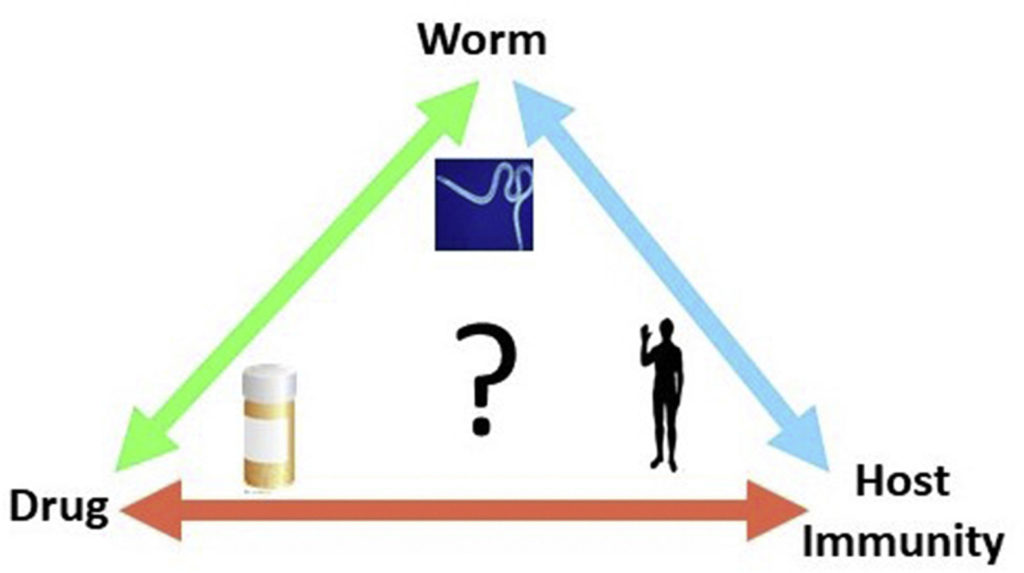
How does building a database fight disease? Data help researchers construct and test their ideas about how to create treatments for diseases or map out ways to halt their spread.
“We don’t give them answers,” Kissinger says. “We give them a framework in which to generate and test hypotheses.”
Kissinger and her team have built databases to take on malaria and other infectious diseases such as toxoplasmosis, cryptosporidiosis and trypanosomiasis. They are also creating tools for studying childhood malnutrition and factors related to disease, and making them accessible to all as they become publicly available. These databases collectively service more than 70,000 unique users a month from more than 100 countries.
To put it simply, her work saves time. It speeds the pace discovery for the next possible solution, the next cure. Without these databases, researchers could spend weeks, months, even years researching existing literature on a disease in the library or recreating work in the lab.

These are tools by biologist for biologists … I think it is that sense of being a member of that community, having your finger on the pulse of what’s going on, that allows you to keep the tools useful. ~ Jessie Kissinger Director, Institute of Bioinformatics
Career Evolution
Kissinger didn’t set out to build databases.
She was trained as a molecular evolutionary biologist, not a computer scientist. “I like to see how molecules change over time,” she says. “When I started in school it was about how a gene or protein evolved.”
It turned out that her field was evolving too. Technology was allowing researchers to understand molecules through bigger data sets. Now, scientists aren’t just looking at individual genes but entire genomes, which are the complete sets of genes in a cell or organism.
As the field evolved, Kissinger learned and embraced the technology. Over time, she shifted her balance away from the so called “wet lab,” where she worked directly with the organisms, to focus mostly on the computer-based “dry lab.”
Her database work started with malaria and continued to expand.
“Now we make 10 different component databases for over 300 organisms (EuPathDB.org), a comparative database to see how conserved genes are across organisms and a new epidemiology database to study the prevalence, spread and factors related to disease in humans (ClinEpiDB.org).” she says.
Kissinger’s team relies on an expert advisory board that helps the researchers customize the databases for each disease community, so they have the largest impact on research. It helps that Kissinger started in a wet lab before diving into informatics.
“These are tools by biologist for biologists,” she says. “We have a lot of computer scientists in the middle, but I think it is that sense of being a member of that community, having your finger on the pulse of what’s going on, that allows you to keep the tools useful.”
SUPPORT OUR RESEARCH
Give to the Center Tropical & Emerging Global Diseases General Fund
[button size=’large’ style=” text=’Give Now’ icon=” icon_color=” link=’https://ctegd.uga.edu/give/’ target=’_self’ color=” hover_color=” border_color=” hover_border_color=” background_color=’#b80d32′ hover_background_color=” font_style=” font_weight=” text_align=’center’ margin=”]
Originally published at https://greatcommitments.uga.edu/story/data-boost/