A CRISPR/Cas9-riboswitch-Based Method for Downregulation of Gene Expression in Trypanosoma cruzi
Few genetic tools were available to work with Trypanosoma cruzi until the recent introduction of the CRISPR/Cas9 technique for gene knockout, gene knock-in, gene complementation, and endogenous gene tagging. Riboswitches are naturally occurring self-cleaving RNAs (ribozymes) that can be ligand-activated. Results from our laboratory recently demonstrated the usefulness of the glmS ribozyme from Bacillus subtilis, which has been shown to control reporter gene expression in response to exogenous glucosamine, for gene silencing in Trypanosoma brucei. In this work we used the CRISPR/Cas9 system for endogenously tagging T. cruzi glycoprotein 72 (TcGP72) and vacuolar proton pyrophosphatase (TcVP1) with the active (glmS) or inactive (M9) ribozyme. Gene tagging was confirmed by PCR and protein downregulation was verified by western blot analyses. Further phenotypic characterization was performed by immunofluorescence analysis and quantification of growth in vitro. Our results indicate that the method was successful in silencing the expression of both genes without the need of glucosamine in the medium, suggesting that T. cruzi produces enough levels of endogenous glucosamine 6-phosphate to stimulate the glmS ribozyme activity under normal growth conditions. This method could be useful to obtain knockdowns of essential genes in T. cruzi and to validate potential drug targets in this parasite.
Noelia Lander, Teresa Cruz-Bustos, and Roberto Docampo. Front Cell Infect Microbiol. 2020 Feb 27;10:68. doi: 10.3389/fcimb.2020.00068. eCollection 2020.

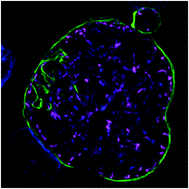 Advanced cell culture methods for modeling organ-level structure have been demonstrated to replicate in vivo conditions more accurately than traditional in vitro cell culture. Given that the liver is particularly important to human health, several advanced culture methods have been developed to experiment with liver disease states, including infection with Plasmodium parasites, the causative agent of malaria. These models have demonstrated that intrahepatic parasites require functionally stable hepatocytes to thrive and robust characterization of the parasite populations’ response to investigational therapies is dependent on high-content and high-resolution imaging (HC/RI). We previously reported abiotic confinement extends the functional longevity of primary hepatocytes in a microfluidic platform and set out to instill confinement in a microtiter plate platform while maintaining optical accessibility for HC/RI; with an end-goal of producing an improved P. vivax liver stage culture model. We developed a novel fabrication process in which a PDMS soft mold embosses hepatocyte-confining microfeatures into polystyrene, resulting in microfeature-based hepatocyte confinement (μHEP) slides and plates. Our process was optimized to form both microfeatures and culture wells in a single embossing step, resulting in a 100 μm-thick bottom ideal for HC/RI, and was found inexpensively amendable to microfeature design changes. Microfeatures improved intrahepatic parasite infection rates and μHEP systems were used to reconfirm the activity of reference antimalarials in phenotypic dose-response assays. RNAseq of hepatocytes in μHEP systems demonstrated microfeatures sustain hepatic differentiation and function, suggesting broader utility for preclinical hepatic assays; while our tailorable embossing process could be repurposed for developing additional organ models.
Advanced cell culture methods for modeling organ-level structure have been demonstrated to replicate in vivo conditions more accurately than traditional in vitro cell culture. Given that the liver is particularly important to human health, several advanced culture methods have been developed to experiment with liver disease states, including infection with Plasmodium parasites, the causative agent of malaria. These models have demonstrated that intrahepatic parasites require functionally stable hepatocytes to thrive and robust characterization of the parasite populations’ response to investigational therapies is dependent on high-content and high-resolution imaging (HC/RI). We previously reported abiotic confinement extends the functional longevity of primary hepatocytes in a microfluidic platform and set out to instill confinement in a microtiter plate platform while maintaining optical accessibility for HC/RI; with an end-goal of producing an improved P. vivax liver stage culture model. We developed a novel fabrication process in which a PDMS soft mold embosses hepatocyte-confining microfeatures into polystyrene, resulting in microfeature-based hepatocyte confinement (μHEP) slides and plates. Our process was optimized to form both microfeatures and culture wells in a single embossing step, resulting in a 100 μm-thick bottom ideal for HC/RI, and was found inexpensively amendable to microfeature design changes. Microfeatures improved intrahepatic parasite infection rates and μHEP systems were used to reconfirm the activity of reference antimalarials in phenotypic dose-response assays. RNAseq of hepatocytes in μHEP systems demonstrated microfeatures sustain hepatic differentiation and function, suggesting broader utility for preclinical hepatic assays; while our tailorable embossing process could be repurposed for developing additional organ models.