It all began with a simple phone call and now, more than a decade later, the Schistosomiasis Consortium for Operational Control and Evaluation (SCORE) is preparing to pass the baton to new groups of investigators working on understanding and controlling schistosomiasis. Under the direction of Dan Colley, a member of the Center for Tropical and Emerging Global Diseases and professor emeritus of microbiology at the University of Georgia, SCORE has advanced the scientific understanding of how to control schistosomiasis and has moved us closer to the elimination of this devastating and sometimes deadly disease.
The beginning
In January 2008, Colley received a phone call from Dr. Julie Jacobson of the Bill & Melinda Gates Foundation (BMGF). She wanted Colley to lead a program on how best to control and move towards elimination of schistosomiasis. This program would not be your typical research program.
First, it would involve national Neglected Tropical Disease (NTD) programs, the World Health Organization (WHO), and others around the globe who were pursuing ways to control morbidity due to Schistosoma mansoni and S. haematobium and their transmission.
“While there were and are now many individual research programs in academia, governments and NGOs working on how to control and eliminate schistosomiasis, SCORE was somewhat different in its size and complexity, allowing it to mount large-scale studies and do so comparatively across different countries,” said Colley.
Second, the program would not focus on basic research, but the results of their research would be more directly applicable to improving national NTD control programs.
“Julie made it clear that the BMGF was not interested, at this time, in funding basic research on either anti-schistosomal drug development or anti-schistosomal vaccine development,” said Colley.
While there are several Schistosoma spp. that infect humans worldwide, this program would focus only on two species, S. mansoni and S. haematobium. Furthermore, they would focus primarily on interventions to control these infections in Africa.
With the parameters laid out and the definition of what constitutes operational research, Colley agreed to gather together a consortium of scientists and SCORE was born: their mission – to undertake operational research that could support National NTD managers in making decisions about how to best control and/or eliminate schistosomiasis in their countries.
“The vision was that research findings would be useful to the WHO in revising current and developing new guidelines on how best to control and move towards elimination of schistosomiasis,” said Colley.
Over the past decade, their mission has not changed but how they pursued that mission evolved as situations change., such as the increased availability of praziquantel, made possible through a donation of the drug by Merck, AG; a more realistic vision of the integration of NTD programs; and the desire of funders to move more quickly from control of morbidity to elimination of the disease.
The projects
 Over the past decade, SCORE has pursued a number of field and laboratory-based projects in 9 African countries. Several of these studies were included in the categories of gaining control, sustaining control of schistosomiasis, or eliminating its transmission.
Over the past decade, SCORE has pursued a number of field and laboratory-based projects in 9 African countries. Several of these studies were included in the categories of gaining control, sustaining control of schistosomiasis, or eliminating its transmission.
The control projects compared how best to deliver Mass Drug Administration (MDA) of praziquantel. Gaining control projects were focused on areas with high prevalence of infection and included research related to subtle morbidity, snail infection, and schistosome population genetics. Sustaining control project focused on areas with moderate prevalence as these areas had already achieved a level of control or simply had lower levels of prevalence.
The original focus of the elimination research project was on Zanzibar and the elimination of S. haematobium. They hoped to inform effective strategies of moving an area of low infection prevalence to total elimination of schistosomiasis. In 2013, with supplemental funding, SCORE expanded its focus on elimination research to focus on S. mansoni in Africa. However, due to conflicts within the country where this research was implemented, SCORE and partners had to withdraw. Instead, SCORE identified another elimination priority on the impact of timed interventions on seasonally transmitted schistosomiasis. This research was conducted in Cote d’Ivoire in a large area with S. haematobium.
Another group of projects focused on tools needed to evaluate control and elimination efforts. They were able to field evaluate the point-of-care circulating cathodic antigen (POC-CCA) urine assay for its use as a mapping tool for S. mansoni infection in humans. They also conducted research and evaluation on highly sensitive and specific human diagnostic tests for schistosomiasis and developed and used tools for schistosome population genetics studies.
Keeping with their mission of supporting national NTD program managers in decision-making, SCORE provided critical information by analyzing and synthesizing existing data in a series of 7 Rapid Answers Projects. Each project resulted in a 2 page brochure providing essential information on a topic of interest to program managers. These brochures are available online at https://score.uga.edu/projects/rapid-answers-project/
And finally, in order to optimize the use of the data generated in these large studies, they collected and made them accessible to the scientific community and other stakeholders. SCORE worked with database programmers and statisticians at UGA to integrate the data from various study sites and conduct analyses of combined data, while providing them to all investigators by depositing them in an open database system, ClinEpiDB.
The legacy
In July, the American Journal of Tropical Medicine and Hygiene published a supplement that summarizes many of the activities, lessons learned, and work that still needs to be done in 16 articles. This supplement is introduced with a guest editorial by N. Robert Bergquist.
Briefly the key findings and take away messages are summarized in Table 1 of the article “Contributions of the Schistosomiasis Consortium for Operational Research and Evaluation (SCORE) to Schistosomiasis Control and Eliminations: Key Findings and Messages for Future Goals, Thresholds, and Operational Research” (https://doi.org/10.4269/ajtmh.19-0787).
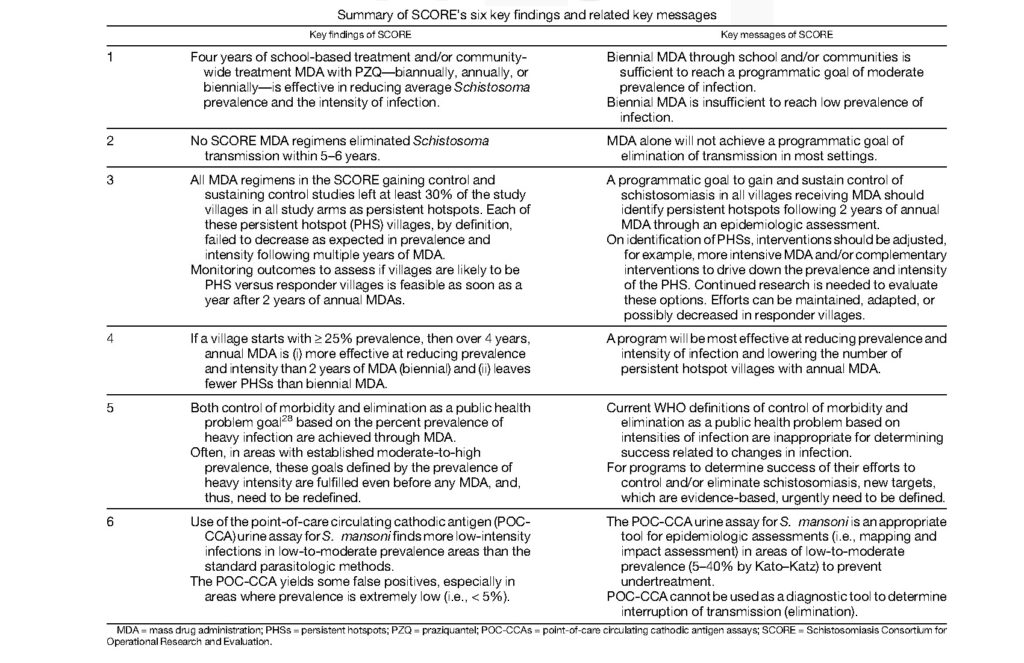
“The impact of SCORE’s findings and messages will depend on their uptake by the WHO/NTD schistosomiasis guidelines development group in the formulation of revised and new guidelines for the control and elimination of schistosomiasis, and then whether national NTD programs consider them worthy of adoption and implementation,” said Colley.
Some of the findings from SCORE are already being implemented, such as the use of the POC-CCA rapid cassette test for mapping S. mansoni prevalence in low-to-moderate areas of schistosomiasis. Other findings, such as the occurrence of persistent hotspots in large-scale MDA programs, are now being considered by other research groups and national NTD programs.
This isn’t really good-bye
SCORE was built on the foundation of a previously BMGF-funded program – the Schistosomiasis Control Initiative (SCI). The goal of SCI, which is now a private non-governmental organization, was to determine if preventive chemotherapy, as recommended by the WHO, could be implemented countrywide to control schistosomiasis.
“It was an ambitious undertaking that SCI accomplished in multiple African countries,” said Colley. “Now knowing that with the funding, persistence, and training, MDA countrywide could be done, the BMGF decided to fund a program to compare the frequency and platforms for MDA distribution.”
Of course, that program became SCORE. They took what SCI learned and asked additional questions about how best to conduct the preventive chemotherapy by MDA, and explore what tools were needed to do it better. Now that SCORE has fulfilled its mission, they are passing their findings and lessons learned, along with recommendations, to other groups. One of these, the recently established Global Schistosomiasis Alliance, which includes some of the same people involved with SCORE, has taken up the baton to help harmonize the continued fight to end schistosomiasis.