In Vitro Diagnostic Assay to Detect SARS-CoV-2-Neutralizing Antibody in Patient Sera Using Engineered ACE-2 Mini-Protein
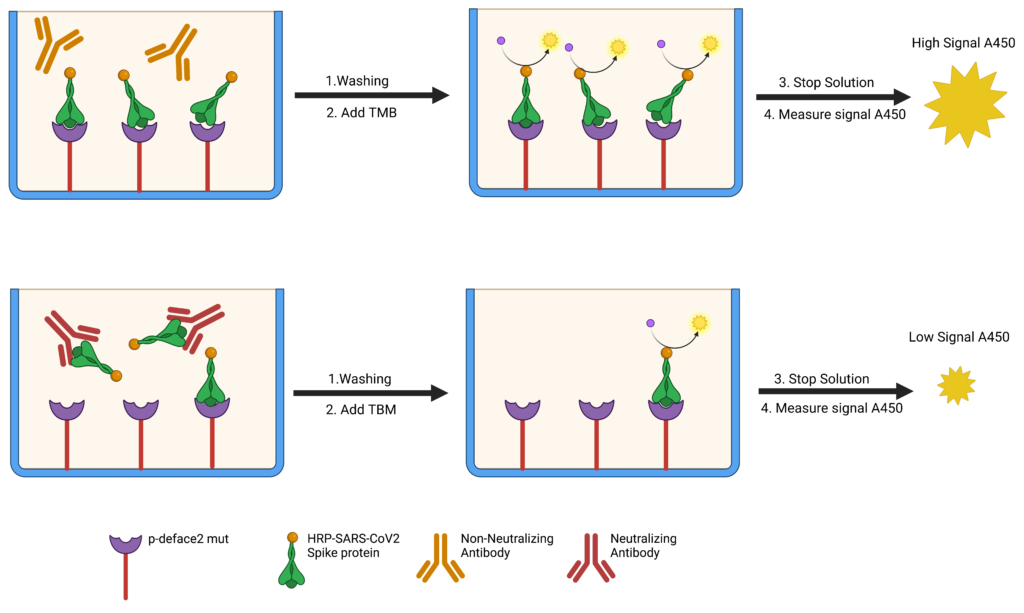 The recent development and mass administration of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) vaccines allowed for disease control, reducing hospitalizations and mortality. Most of these vaccines target the SARS-CoV-2 Spike (S) protein antigens, culminating with the production of neutralizing antibodies (NAbs) that disrupt the attachment of the virus to ACE2 receptors on the host cells. However, several studies demonstrated that the NAbs typically rise within a few weeks after vaccination but quickly reduce months later. Thus, multiple booster administration is recommended, leading to vaccination hesitancy in many populations. Detecting serum anti-SARS-CoV-2 NAbs can instruct patients and healthcare providers on correct booster strategies. Several in vitro diagnostics kits are available; however, their high cost impairs the mass NAbs diagnostic testing. Recently, we engineered an ACE2 mimetic that interacts with the Receptor Binding Domain (RBD) of the SARS-2 S protein. Here we present the use of this engineered mini-protein (p-deface2 mut) to develop a detection assay to measure NAbs in patient sera using a competitive ELISA assay. Serum samples from twenty-one patients were tested. Nine samples (42.8%) tested positive, and twelve (57.1%) tested negative for neutralizing sera. The data correlated with the result from the standard commercial assay that uses human ACE2 protein. This confirmed that p-deface2 mut could replace human ACE2 in ELISA assays. Using bacterially expressed p-deface2 mut protein is cost-effective and may allow mass SARS-CoV-2 NAbs detection, especially in low-income countries where economical diagnostic testing is crucial. Such information will help providers decide when a booster is required, reducing risks of reinfection and preventing the administration before it is medically necessary.
The recent development and mass administration of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) vaccines allowed for disease control, reducing hospitalizations and mortality. Most of these vaccines target the SARS-CoV-2 Spike (S) protein antigens, culminating with the production of neutralizing antibodies (NAbs) that disrupt the attachment of the virus to ACE2 receptors on the host cells. However, several studies demonstrated that the NAbs typically rise within a few weeks after vaccination but quickly reduce months later. Thus, multiple booster administration is recommended, leading to vaccination hesitancy in many populations. Detecting serum anti-SARS-CoV-2 NAbs can instruct patients and healthcare providers on correct booster strategies. Several in vitro diagnostics kits are available; however, their high cost impairs the mass NAbs diagnostic testing. Recently, we engineered an ACE2 mimetic that interacts with the Receptor Binding Domain (RBD) of the SARS-2 S protein. Here we present the use of this engineered mini-protein (p-deface2 mut) to develop a detection assay to measure NAbs in patient sera using a competitive ELISA assay. Serum samples from twenty-one patients were tested. Nine samples (42.8%) tested positive, and twelve (57.1%) tested negative for neutralizing sera. The data correlated with the result from the standard commercial assay that uses human ACE2 protein. This confirmed that p-deface2 mut could replace human ACE2 in ELISA assays. Using bacterially expressed p-deface2 mut protein is cost-effective and may allow mass SARS-CoV-2 NAbs detection, especially in low-income countries where economical diagnostic testing is crucial. Such information will help providers decide when a booster is required, reducing risks of reinfection and preventing the administration before it is medically necessary.
Bruna Andersen Pereira de Jesus, Anderson Albino Gomes, Alex E Clark, Tayse Andrade Rodrigues, Melissa Ledgerwood-Lee, Westley Van Zant, Howard Brickner, Meiqiao Wang, David L Blum, Maria B Cassera, Aaron F Carlin, Eliah S Aronoff-Spencer, Gustavo Felippe da Silva, Maria de Lourdes Borba Magalhães, Partha Ray. Viruses. 2022 Dec 18;14(12):2823. doi: 10.3390/v14122823.


